Fast_ep
At the end of fast_dp two additional pipelines are run, an automated experimental phasing system fast_ep and dimple, a difference map pipeline. These are to answer the questions “can I determine my heavy atom substructure” and “do I have a ligand bound” respectively.
The fast_ep pipeline (Figure 1) first performs an initial analysis to decide if there is an anomalous signal present. If this is the case then SHELXC is run to generate the input files needed for substructure determination with SHELXD. These are then duplicated and modified to generate input files for: all possible (enantiomorph-unique) spacegroups and all sensible numbers of heavy atom sites. Each of these tasks are then run on the cluster using the multi-processor version of SHELXD, and the solution with the highest combined figure-of-merit taken as correct. This solution is then phased in both enantiomorphs and with solvent fractions in the range 25% – 75% in 5% steps. The solution with the highest CC is then taken as correct and converted to MTZ format for viewing in Coot, typically within three minutes (Figure 2).
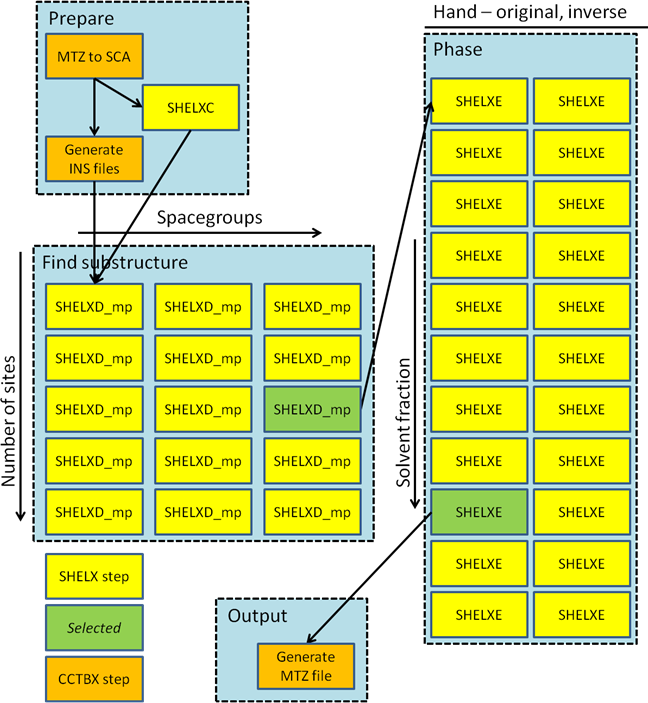
Figure 1 Workflow for experimental phasing with fast_ep
Figure 2 Illustration of results of experimental phasing with fast_ep


 Macromolecular Crystallography
Macromolecular Crystallography
