At the top of the main data view in ISPyB there are short-cut buttons to various useful functions:
![]()
Summaries of all data from a visit can be obtained, which may be useful for comparing different crystals in the same project.
Containers are assigned prior to beamtime to match the puck position in the robot dewar. For remote collections this is normally carried out by the local contact, but should be checked by the user.
The summary link summises Data collections in a tabular format:
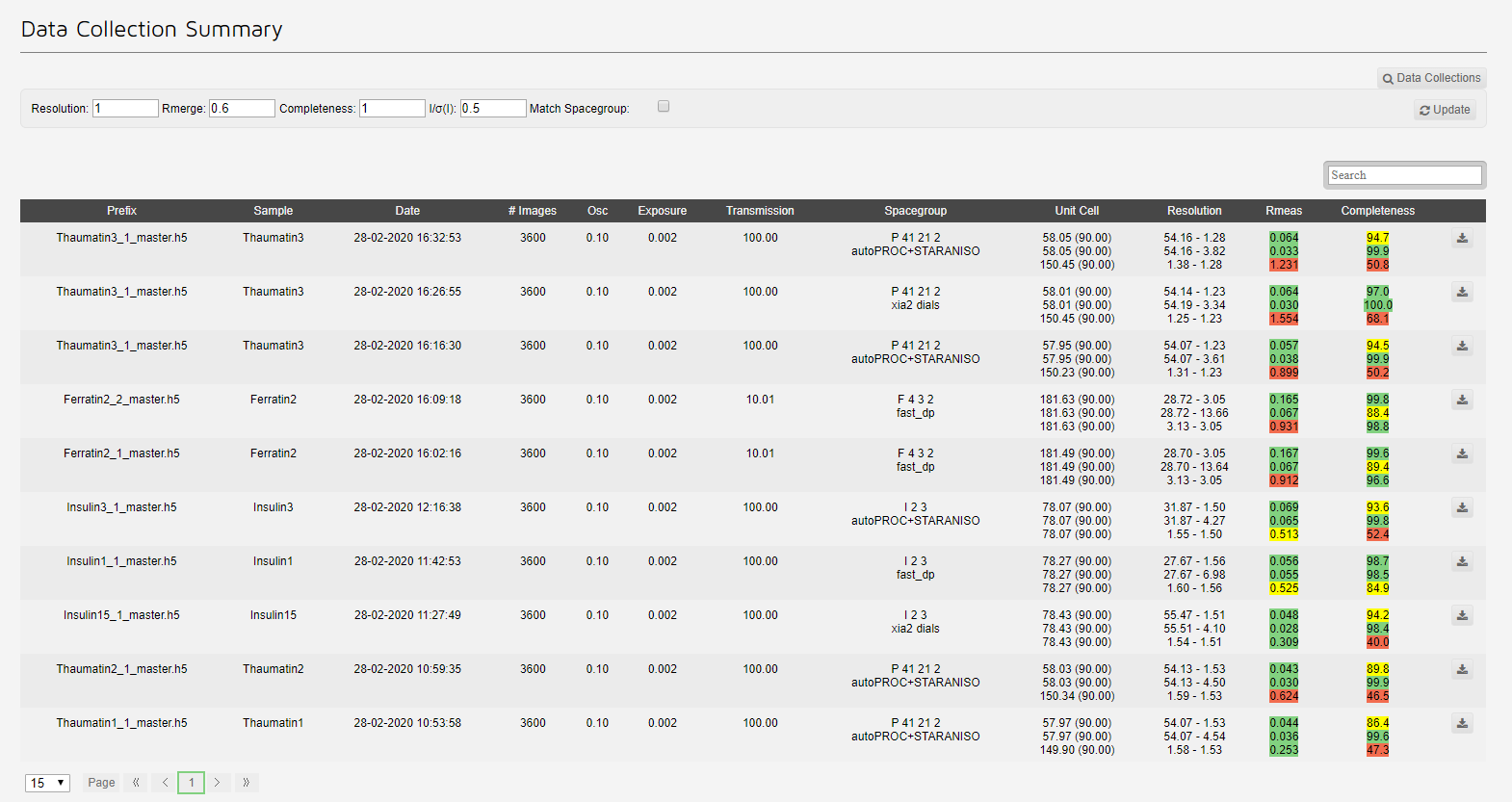
The table contains all data collections, each summarised with:
Note that the data can be filtered using the search box, but the sorting (clicking the top of a column) is currently not working.
The autoprocessing summary shows which autoprocessing pieplines have run:
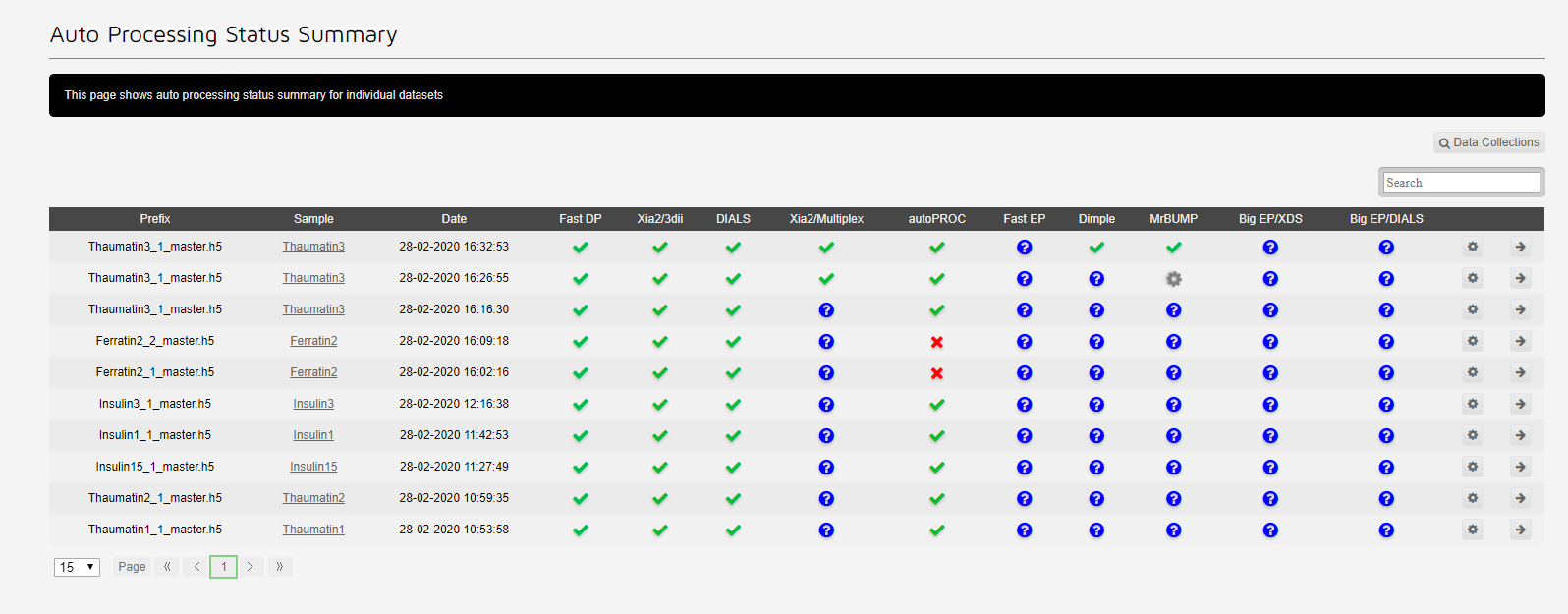
The cog allows reprocessing for the dataset. The arrow redirects to the dataset.
A quick link to the main summary of Visit Statistics which are discussed further here.
Hover over the icon to see a list of people involved in the visit.
Users are added to proposals by the BAG PI, in UAS. You need to be added to a proposal to be able to access data from the visit, as well as those Users involved in data collection.
Hover over the icon to see which Dewars are assigned to this visit.
The sample changer status shows a representation of the pins available for your visit.
You can also reach a quick view of this on the side-bar of an active visit.
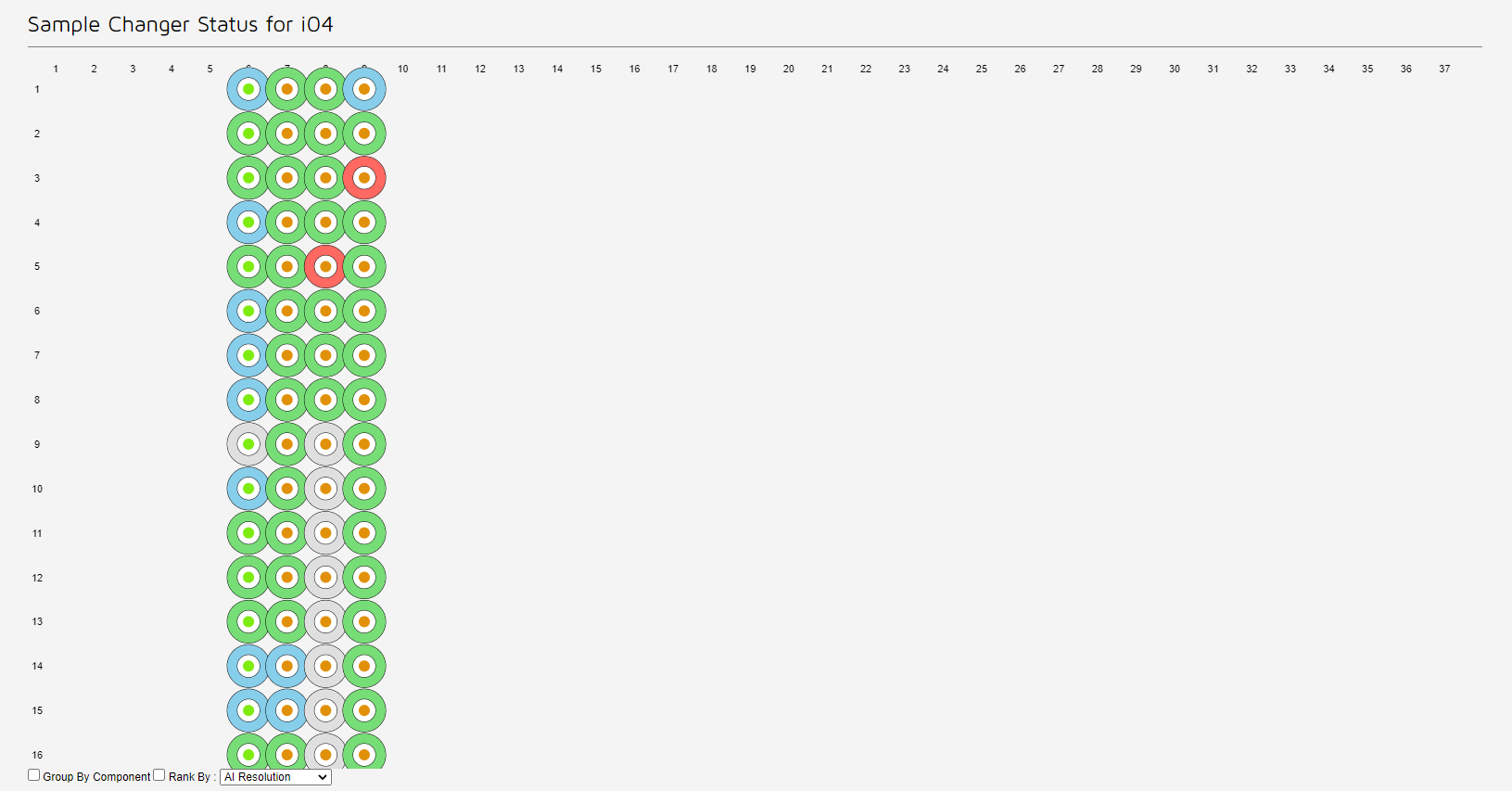
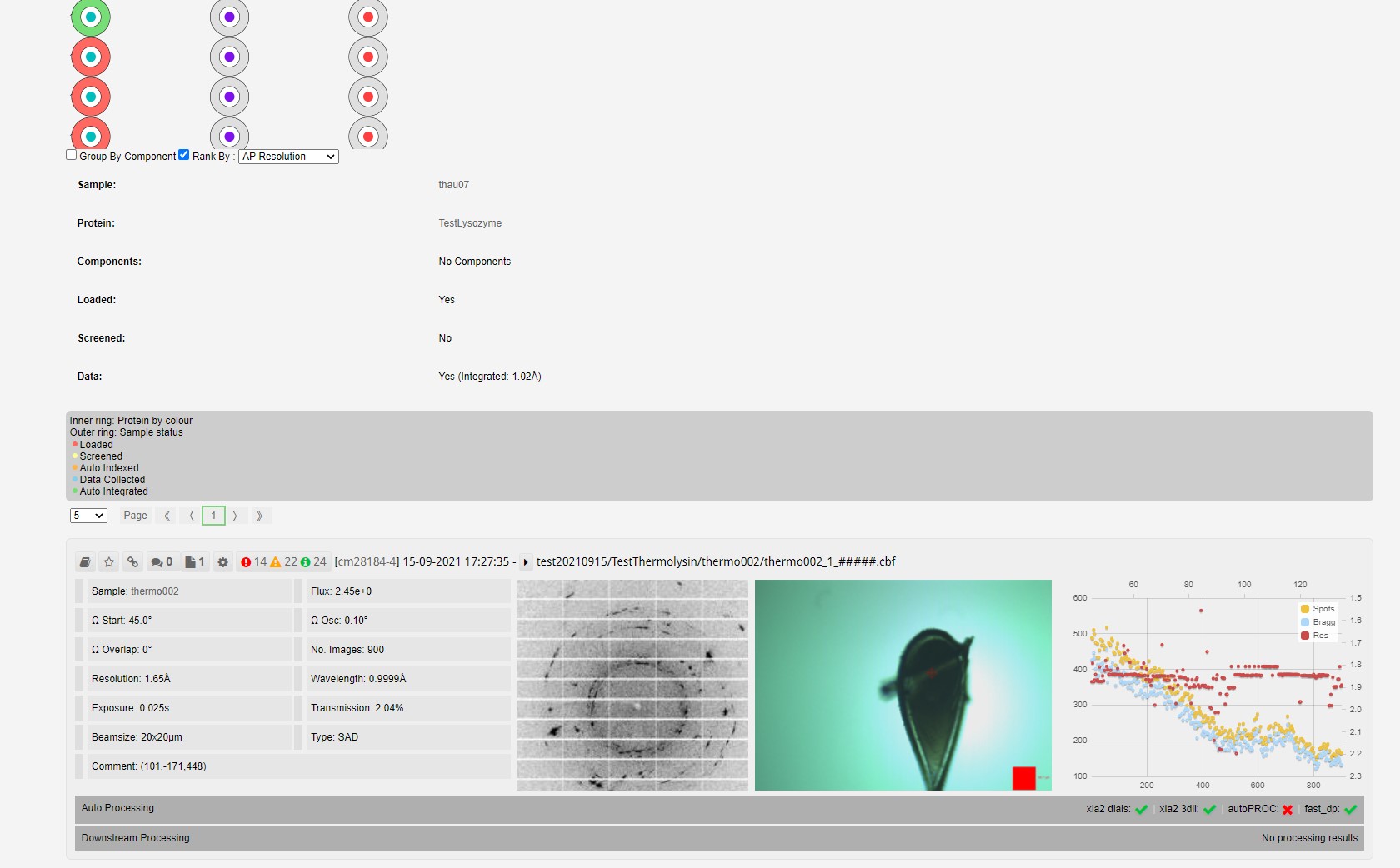
Each sample is represented by a circle, a ring of solid colour surrounding a central dot. In the default view
When hovering over a sample the remainder of those protien samples are highlighted:
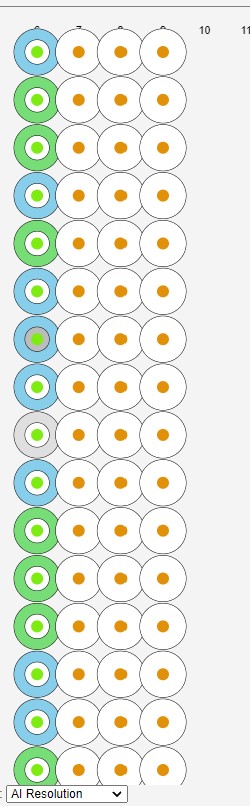
The proteins not highlighted are shown in white.
The filtering per can be used to rank by completeness or resolution, this alters the colours of the ring to the relative resolution or completenes compared to that of the best dataset for that protein. This can be useful on assessing whether any further datasets are required or recollections of exisiting samples need to be made.

If a circle is clicked the data colections and grid scans for that sample are shown below.
A reprocessing panel appears to re-submit automatic processing pipelines with different parameters. See full details in reprocessing data.
This brings you to a quick view which shows the beamline status for your visit, and the webcams.
As most MX visits to Diamond are done by remote access, you will not get the same notifications about beam dumps that you would get if you were a User on site.
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.