To register samples, it is first necessary to create a new shipment, and detail information on all proteins.
Once a Shipment has been created click the "Add Container" button on a dewar in the shipment
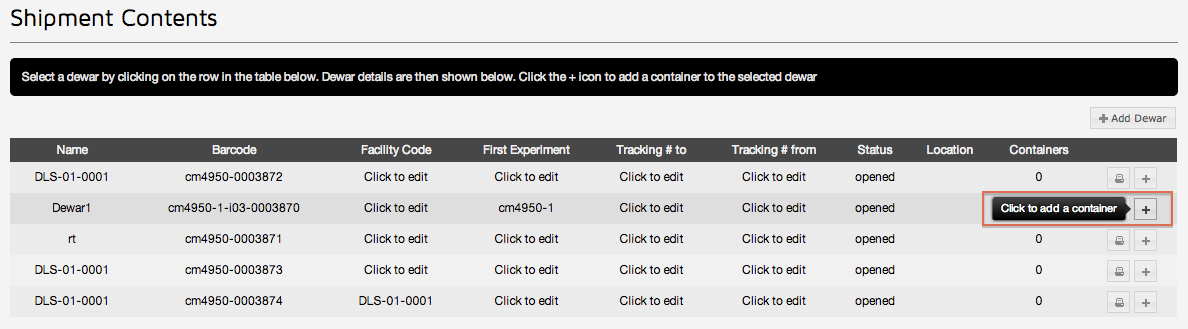
This will take you to the add container page. Here you can register samples for this puck. First select the type of container, which in most cases will be a puck:
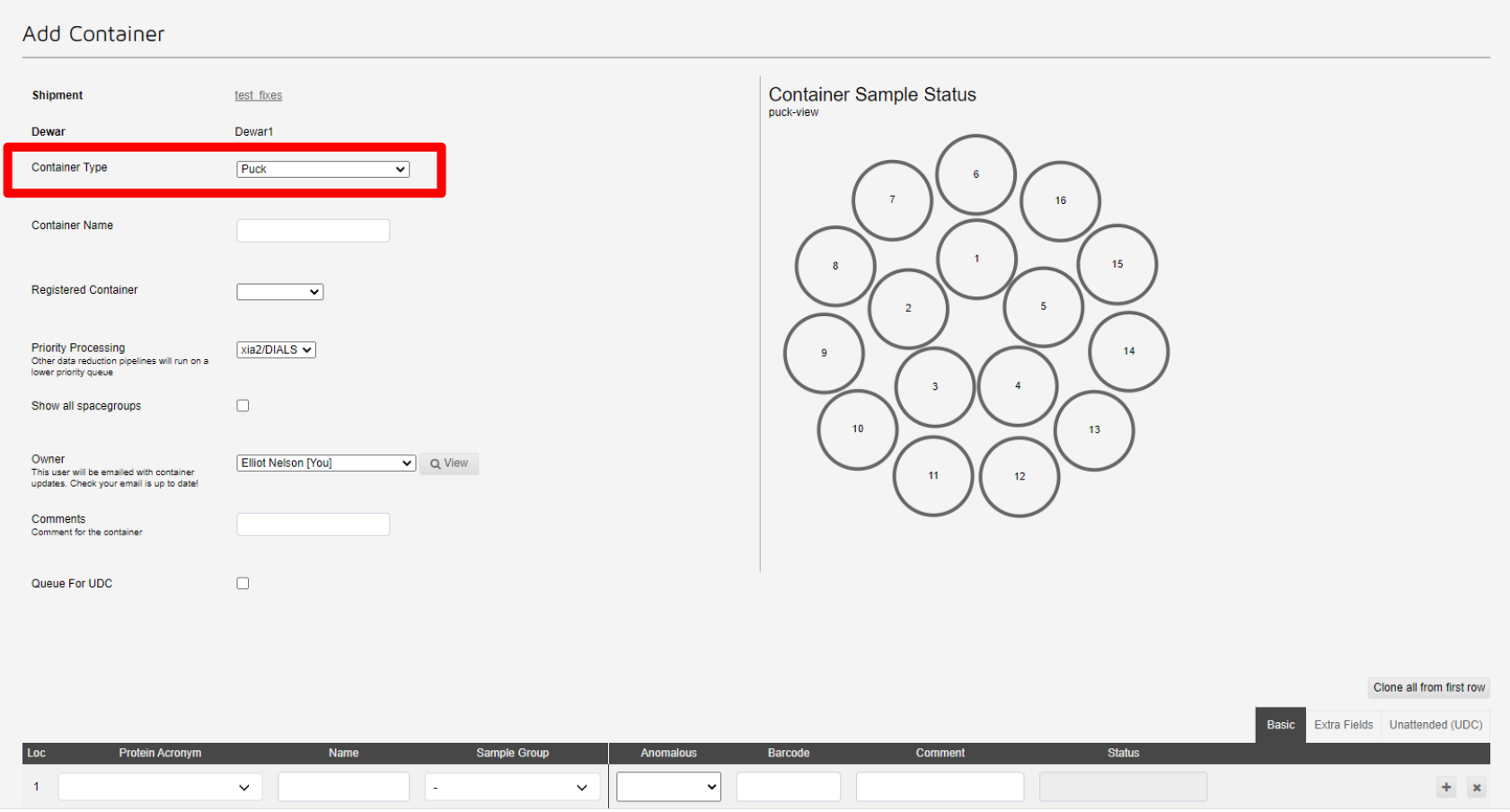
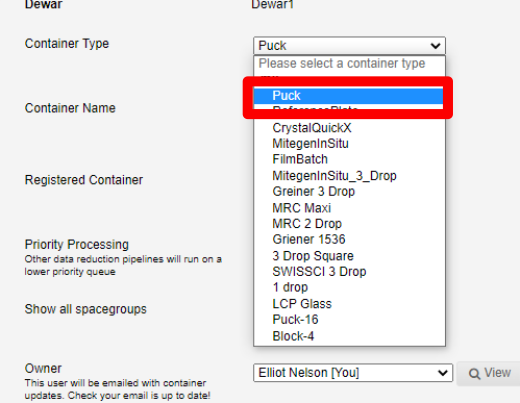
Next select the registered container from the drop-down list:
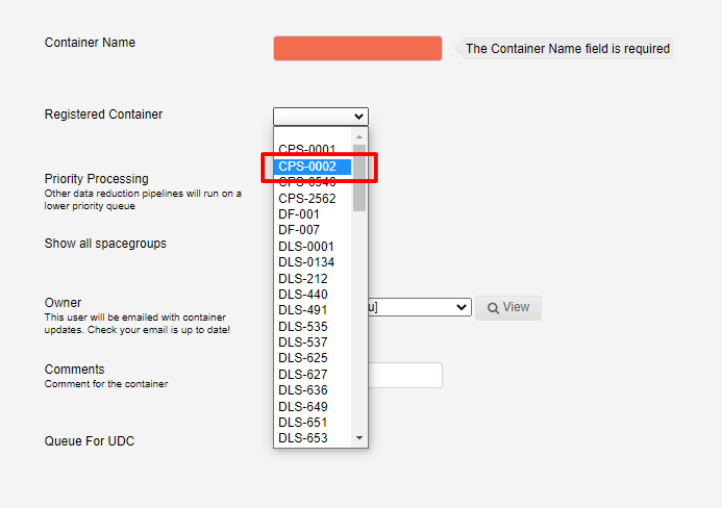
This will autopopulate the container name.
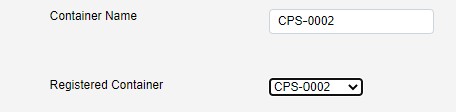
The container name is editable if needed, but it is recommended to just use the registered container name. If you need to register further containers against your proposal, please contact [email protected] for academic proposals, or [email protected] for industrial proposals.
Next you should select a pipeline for priority processing:
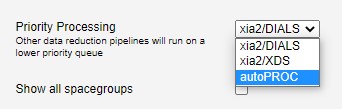
This defines which autoprocessing pipeline is given the highest priority. Lower priority tasks will be run on a lower priority queue, and will usually take longer to complete.
Fast DP is prioritised above other pipelines first to give a quick result for live review of the samples.
Show all spacegroups can be ticked if you wish to provide a space group for the sample, and thus processing pipelines, which is not normally processed for macromolecular crystallography. This may be relevant if you are looking at racemic protein crystallography or chemical crystallography.
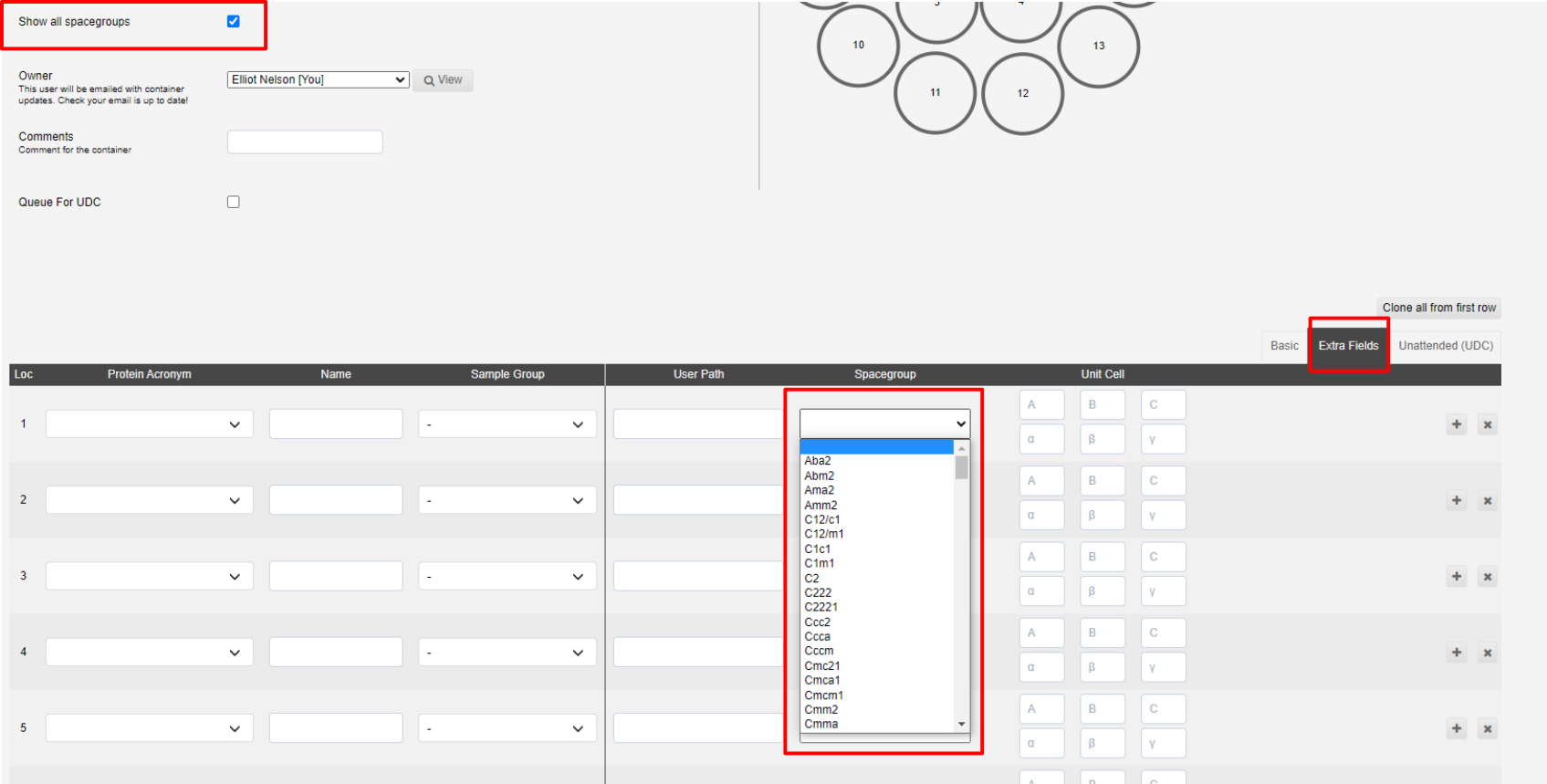
The spacegroups are shown under the extra-fields tab of the sample interface.
The interface for addding samples is split into three tabs; Basic, Extra Fields and Unattended. The minimal information for samples not for unattended data collection is the Protein Acronym and Name:
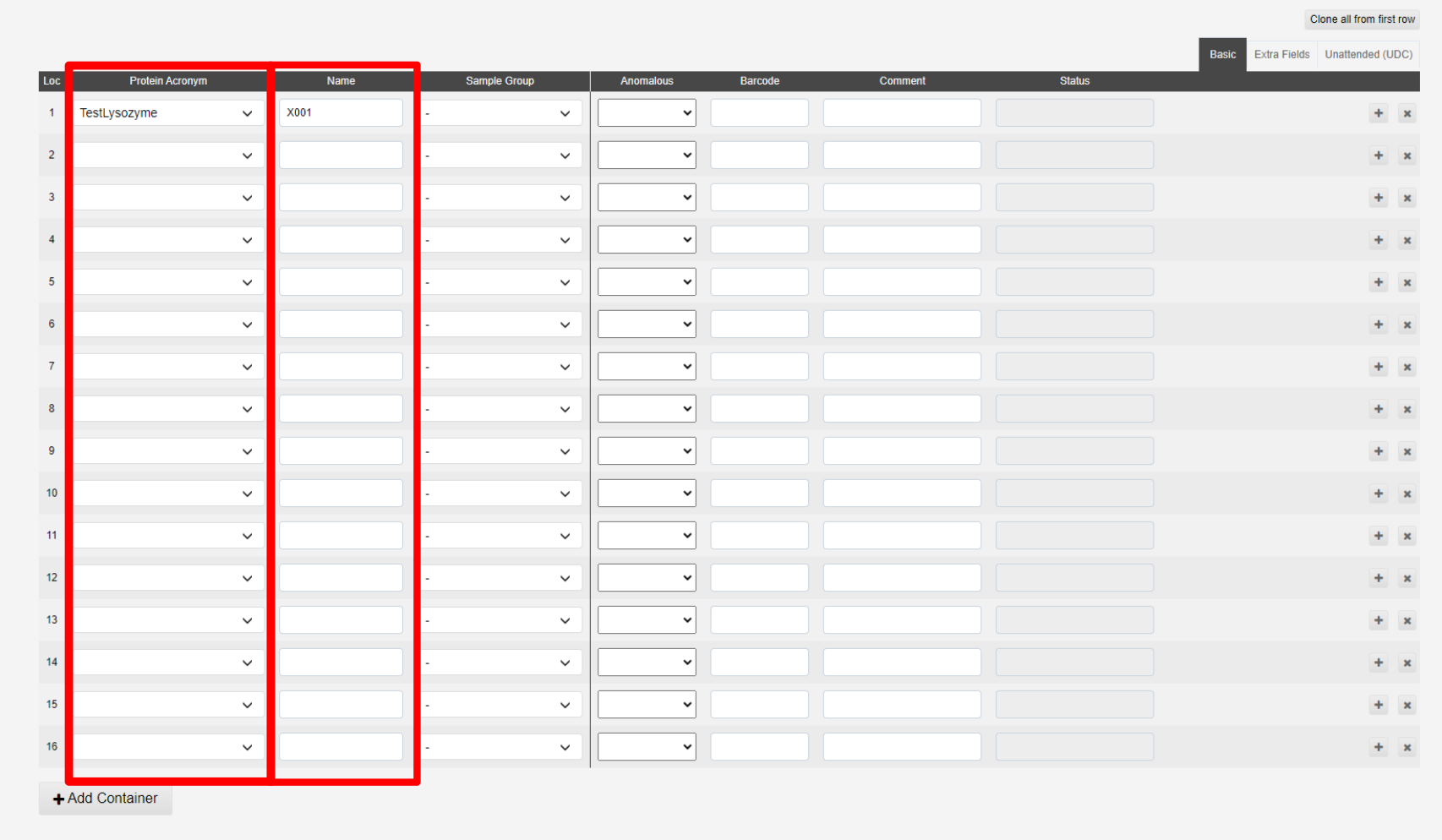
The + button, marked a below, allow you to duplicate the previous sample with a sequential name (i.e. X1 > X2). The x button, marked d below, allows you to remove a sample:
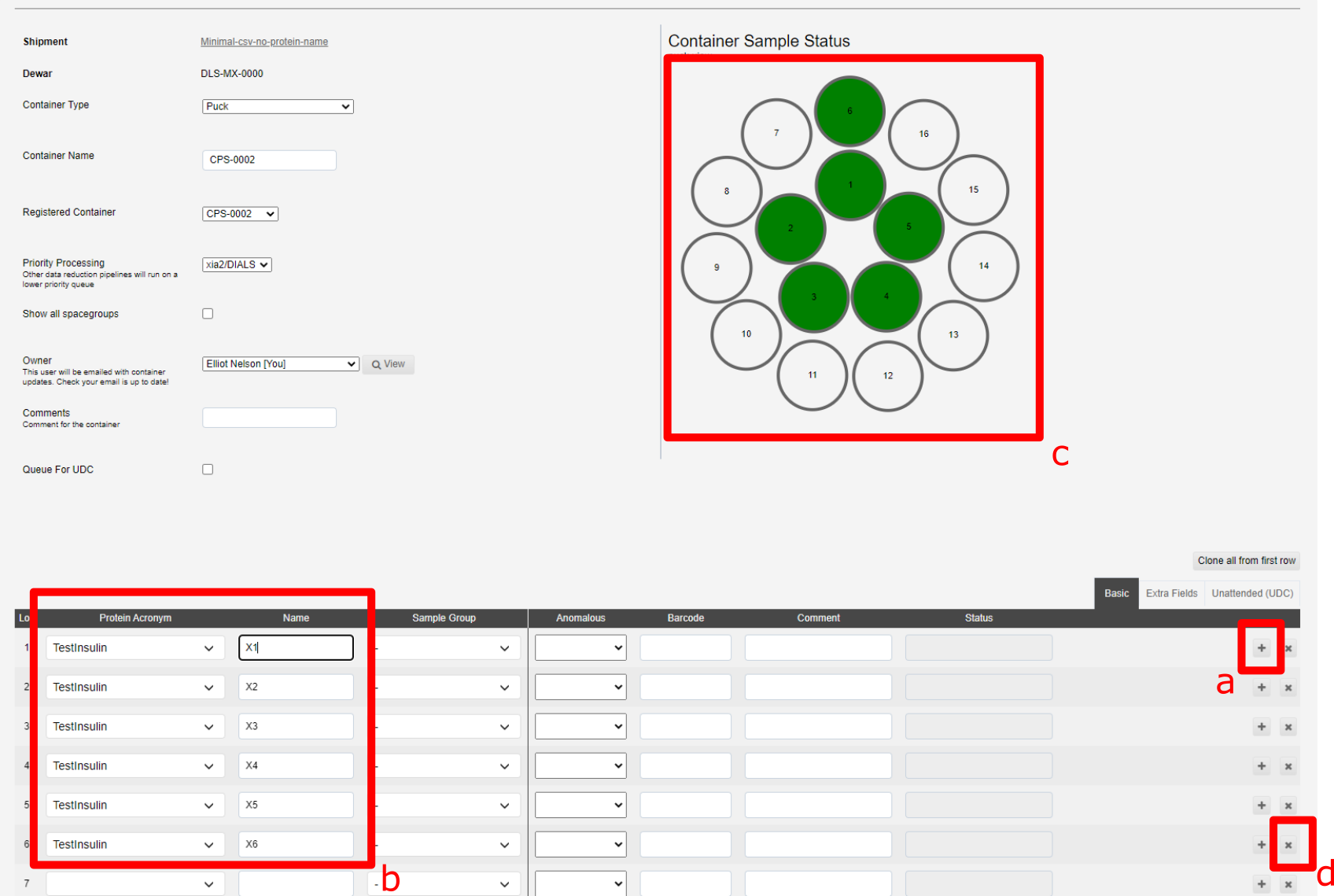
The samples in b are shown in the puck view c above, green filled circles implies sufficent information has been provided.
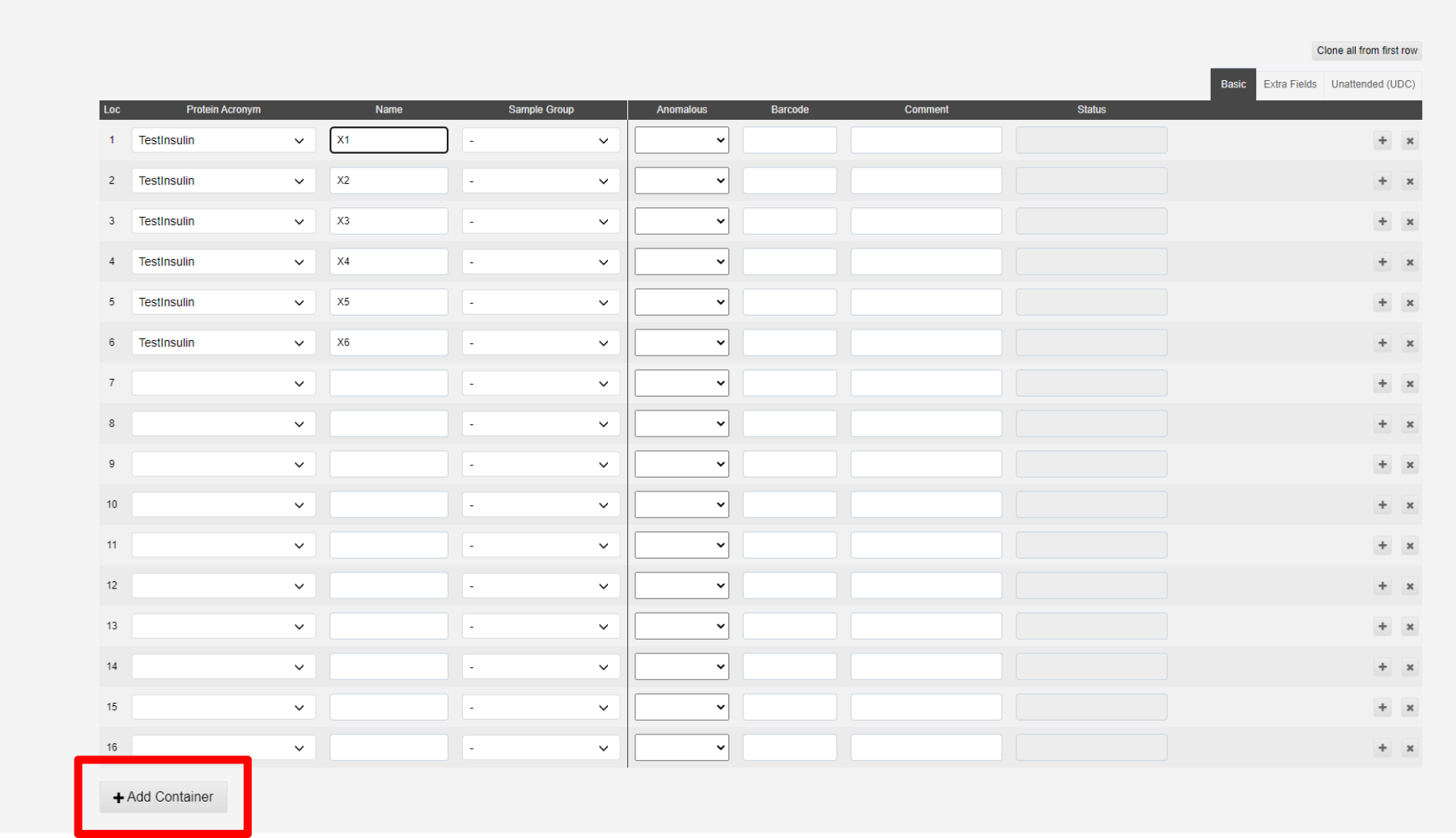
To save your new puck click "Add Container" at the bottom of the page
This will bring up a banner allowing you to see and edit the puck further in the container page.
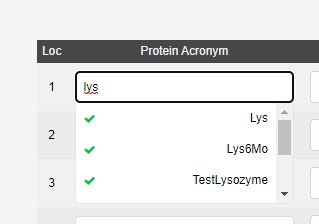
The protein acronym selects from a list of samples approved by Diamond Health and Safety. The samples should be submitted in advance using UAS, as the team have to review the safety information, and then a propagation from UAS to ISPyB runs. If you are unsure why a protein acronym is missing please contact [email protected] for academic proposals, or [email protected] for industrial proposals.
The sample group field is currently used for VMXi and Unattended data collection (Screening).
The barcode of your pin can be provided, a check against the barcode scanned when loading the pin on the goniometer is done within GDA, and reported within ISPyB when a mismatch is found.
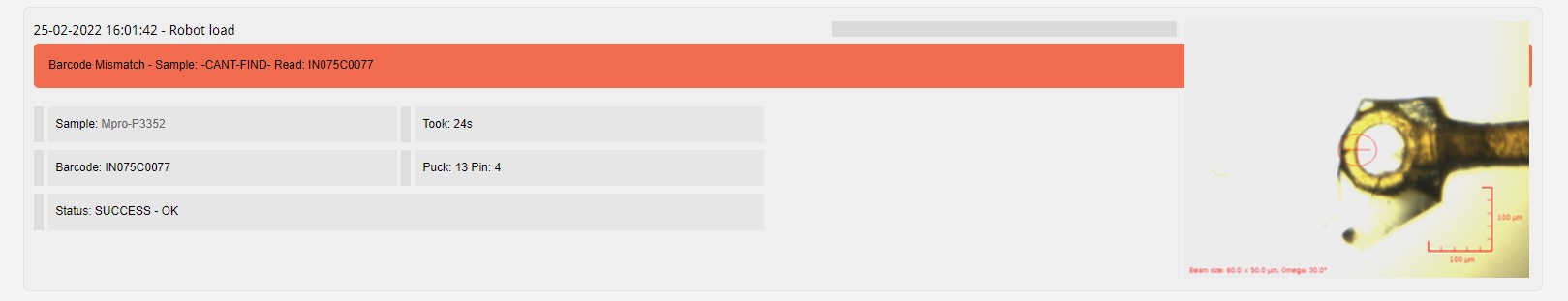
The comment field is a free text field, which you can use to help organise your samples, i.e. Needle like crystal, Ethylene glycol dip soak at 25%.
To run automated phasing pipelines (Fast EP or Big EP) the anomalous scatterer must be supplied in the sample registration.
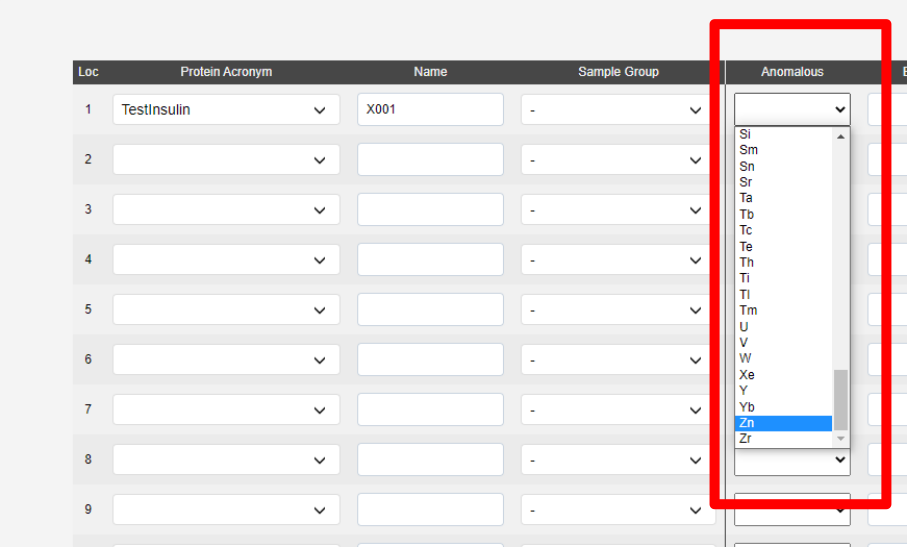
The extra fields tab contains a few optional fields:
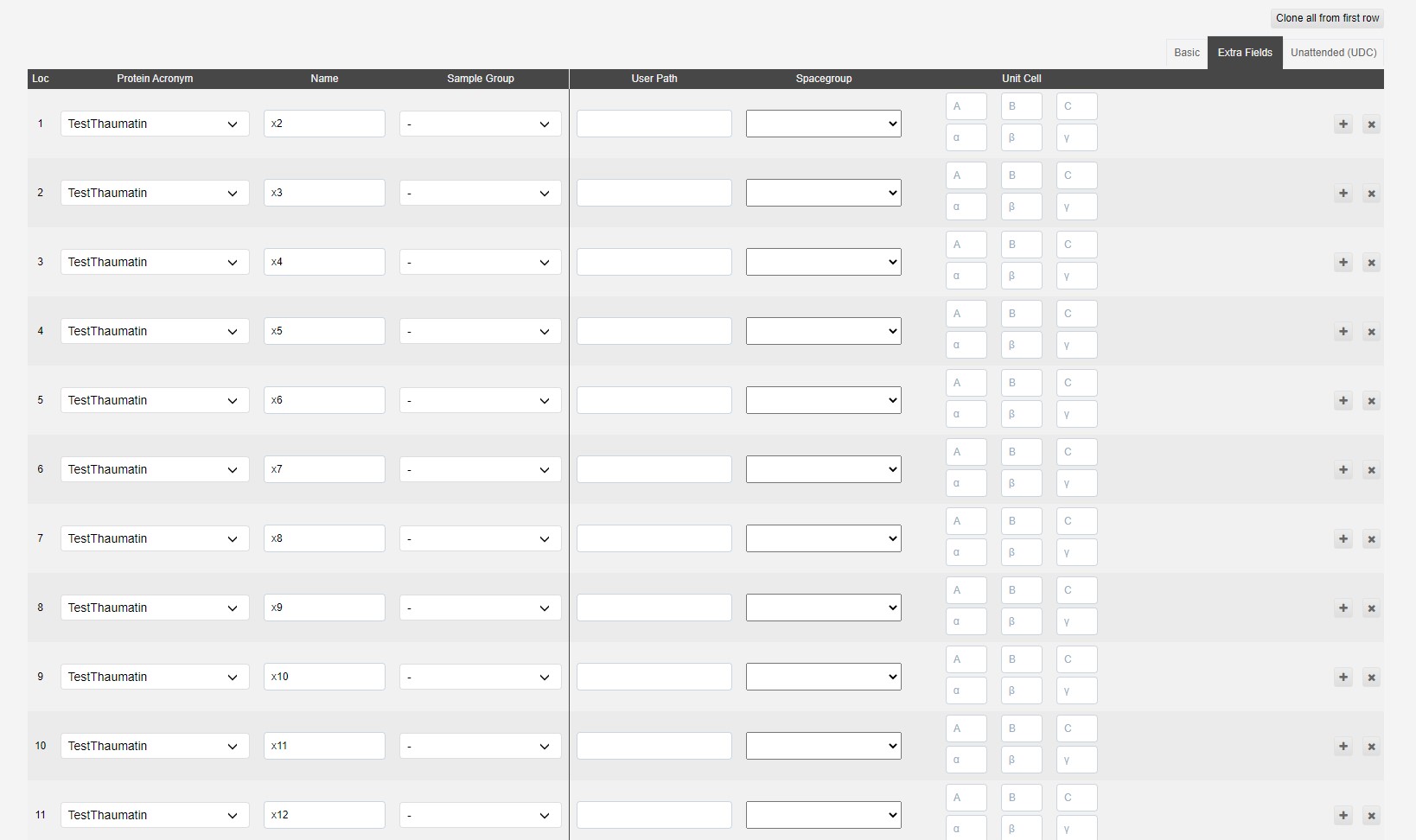
The user path adds an extra folder structure to file path
i.e /dls/i03/data/2021/<visit>/auto/userpath1/userpath2/<protein_acronym>/<sample_name>/
The spacegroup can be provided to trigger autoprocessing assuming that space group, which may be useful to give results in the expected space group, especially when it may be hard to determine automatically. We will also run autoprocessing without knowledge of the space group.
Details on unattended collection are given in Preparation for UDC.
Samples can be cloned from the first line of the table using the Clone from first sample button:
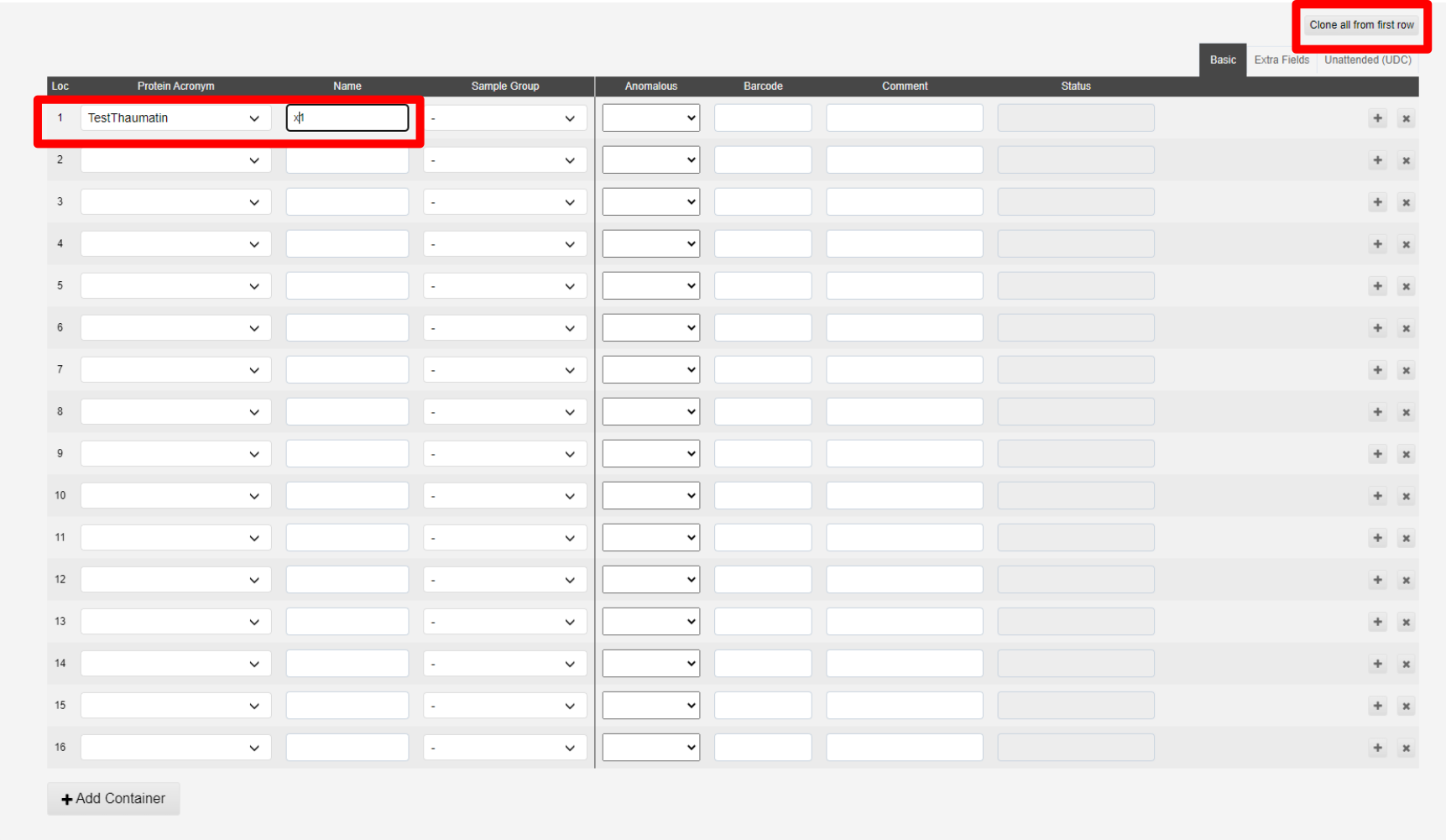
This will populate all spaces in the puck:
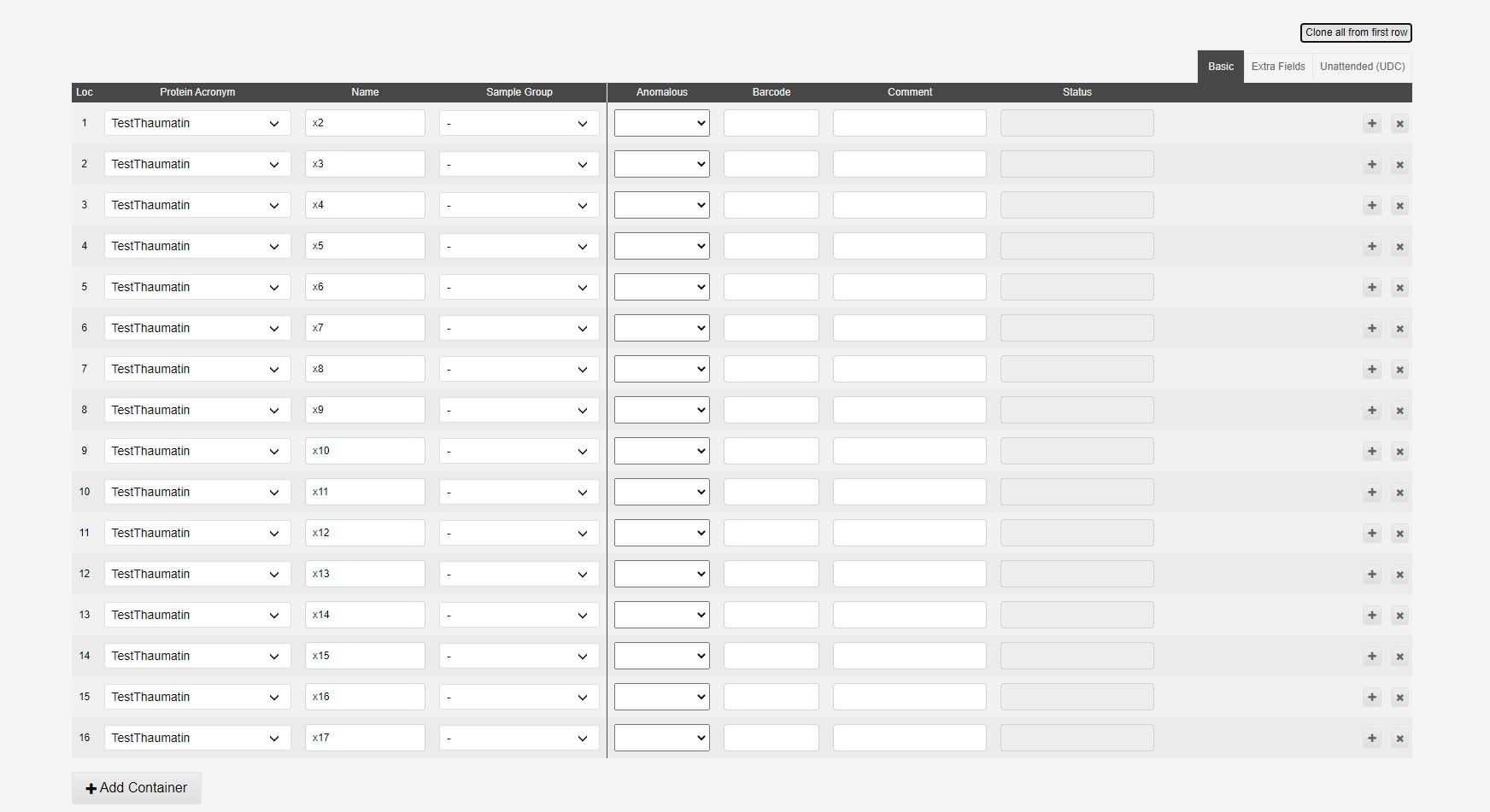
Lines can be deleted or altered to add different samples to a puck.
Errors are reported if the container cannot be submitted:, in a red text box above the Add container button. These diappear live as the error is fixed.
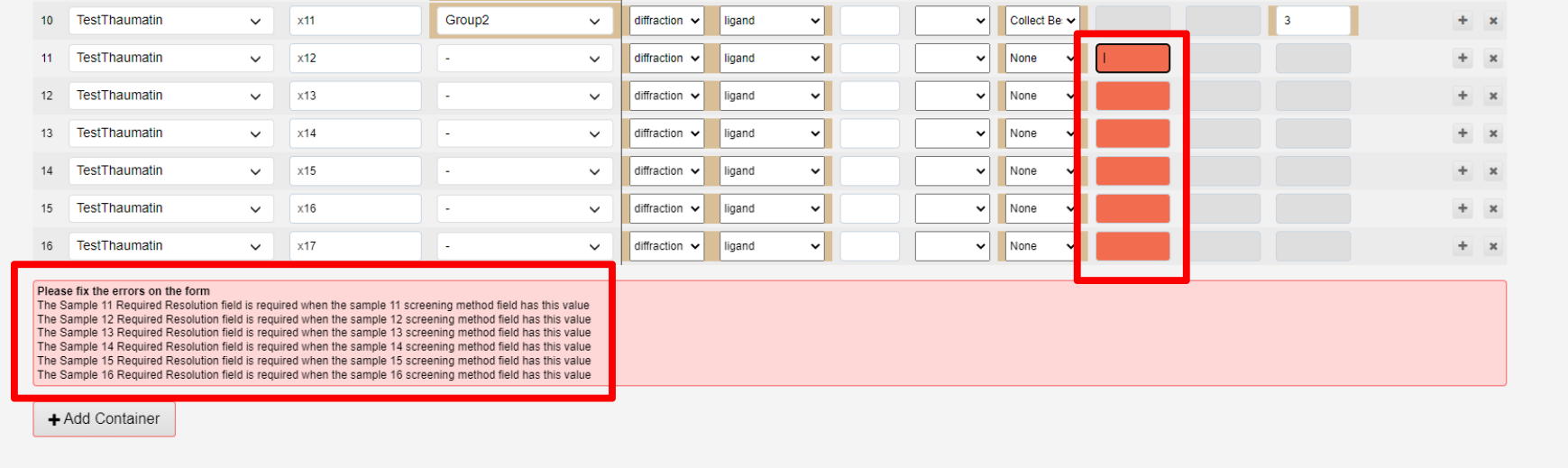
See Uploading samples using CSV file for uploading a larger number of samples.
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.