Uploading Samples using CSV file
This upload mechanism is subject to change, contact [email protected] / [email protected] if you require assistance, or believe this page is outdated.
When dealing with a large number of samples it is possible to use a correctly specified csv file which can then be uploaded using a script. It may be useful to help integrate with your laboratory information management system.
This method for sample upload is suitable for all access modes: Responsive Remote, On site visits, Industrial Mail-in and Unattended Data Collection (UDC).
Steps to complete prior to upload
UAS
As with samples that are registered via ISPyB, you will need to first Ensure sample registered on UAS. Check that the experiment risk assessment (ERA) is validated by Diamond Safety, Health and Environment (SHE) group before preparing the shipment.
The protein acronym to be used for sample upload to ISPyB must exactly match the protein acronym used in the ERA. To define a new protein based on a previously validated sample (e.g. a seleno-methionine derivative or point mutant) the original approved sample can be cloned as described here.
When a sample ERA has been validated, the approved protein acronyms will be transferred to ISPyB. It will only be possible to upload the CSV file after this step is finalised. The transfer runs every 4 hours, so there may be some waiting time before you can upload a newly validated sample.
Creating Shipment
Next you should create a shipment. The shipment name specified in the CSV must be an exact match with the shipment name created via the ISPyB/SynchWeb interface.
Preparing the csv file
Next prepare the csv file. The format of the file is a comma delimited .csv file with up to 29 columns.
Downloadable template csv, with all columns and 1 sample.
Each line (row) in the file represents one sample and each sample must be listed. Data fields (columns) cannot contain commas. The minimal number of columns to be included is 15, so all lines must have at least this many columns, and if any column values are specified for any row, all rows must have at least that many columns specified.
The fields used in the CSV are as below in this order:
| Field | Required? | UDC required? | Description | Example |
| proposalCode | First Line | First Line | Proposal type. i.e. mx, in, sw | mx |
| proposalNumber | First Line | First Line | Proposal number | 23694 |
| visitNumber | Visit number | 72 | ||
| shippingName | First Line | First Line | Name of shipment. Normally shipment should be created in synchweb first. | minimal_csv-2 |
| dewarCode | All Lines | All Lines | Dewar code | DLS-MX-0000 |
| containerCode | All Lines | All Lines | Puck barcode. Needs to match exactly, case sensitive | CPS-0001 |
| preObsResolution | Not used | |||
| minimalResolution | Screening: Better Than | Minimal resolution at which to collect datasets when using the Better Than screening strategy | 2.5 | |
| oscillationRange | Not used | |||
| proteinAcronym | All Lines | All Lines | Protein acronym, must match an approved sample in ISPyB (and thereby UAS). | TestLysozyme |
| proteinName | All Lines | All Lines | Protein name. | TestLysozyme |
| spaceGroup | Space group. | P32 | ||
| sampleBarcode | All Lines | All Lines | Pin barcode. Can be set to any value if the pin is not barcoded. | AB3214 |
| sampleName | All Lines | All Lines | Sample name, should be unique. | x0001 |
| samplePosition | All Lines | All Lines | Position in puck | 1 |
| sampleComments | Comments on sample | |||
| cell_a | Cell dimension a. | 37 | ||
| cell_b | Cell dimension b. | 37 | ||
| cell_c | Cell dimension c. | 73 | ||
| cell_alpha | Cell angle alpha. | 90 | ||
| cell_beta | Cell angle beta. | 90 | ||
| cell gamma | Cell angle gamma. | 90 | ||
| subLocation | Not used | |||
| loopType | Not used | |||
| requiredResolution | All Lines | UDC resolution you expect crystals to diffract to. | 1.8 | |
| centringMethod | All Lines | UDC centring method (diffraction, optical). Diffraction is strongly recommended | diffraction | |
| experimentKind | All Lines | UDC recipe to use (native, phasing, ligand or stepped). See UDC webpages for details | native | |
| radiationSensitivity | Not used | |||
| energy | If needed | UDC energy in electron volts. Only add if energy needs to be specified. See UDC webpages for details | 12700 | |
| userPath | If needed |
Adds user structure to file path. i.e /dls/i03/data/2021/<visit> /auto/userpath1/userpath2... |
||
| screenAndCollectRecipe | Screening | Method to be Used for screening strategy: "all" equivalent to the Better Than strategy, "best" equivalent to Collect Best N strategy, leave blank for no screening strategy | all | |
| screenAndCollectNValue | Screening: | Number of samples to collect when using the Collect best N screening strategy | 3 | |
| sampleGroup | Screening | Sample group that is used to define which crystals are related to one another for the screening strategy | Group_3 |
Upload script from Diamond
Access to diamond file systems can be via No Machine Client, or ssh. File transfer can be done via drag and drop in No Machine, WinSCP, scp, rsync or via a web service open on NX and a local client.
To upload the shipment to a proposal, move the .csv file to one of the supported locations:
- Home directory
- In the tmp folder of the target visit.
- In the tmp folder of a different visit in the same proposal. This can be useful e.g. if the target visit directory doesn't exist yet.
and run the upload csv command:
/dls_sw/apps/ispyb/bin/uploadcsv <Path to CSV FIle>/<csv filname>.csv
Flags are available to alter the behaviour of the upload script:
- --UDC so that the container is queued for UDC. Equivalent to --queuecontainer:
- /dls_sw/apps/ispyb/bin/uploadcsv --UDC <Path to CSV FIle>/<csv filname>.csv
If successful, the command will respond simply with "Done!".
Please ensure that the upload was successful by checking the content of the shipment in ISPyB.
You may see a warning like this, don't worry about it:
- WARNING: Not setting lab contacts for shipment as the csv file owner <fedid> is not a lab contact for proposal <proposal>.
Minimal Working CSV
We first show the upload of a Minimal working example CSV file.
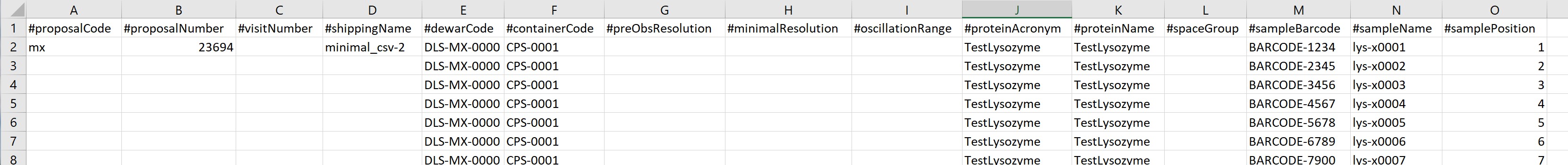
First a shipment is generated in ISPyB/ Synchweb:
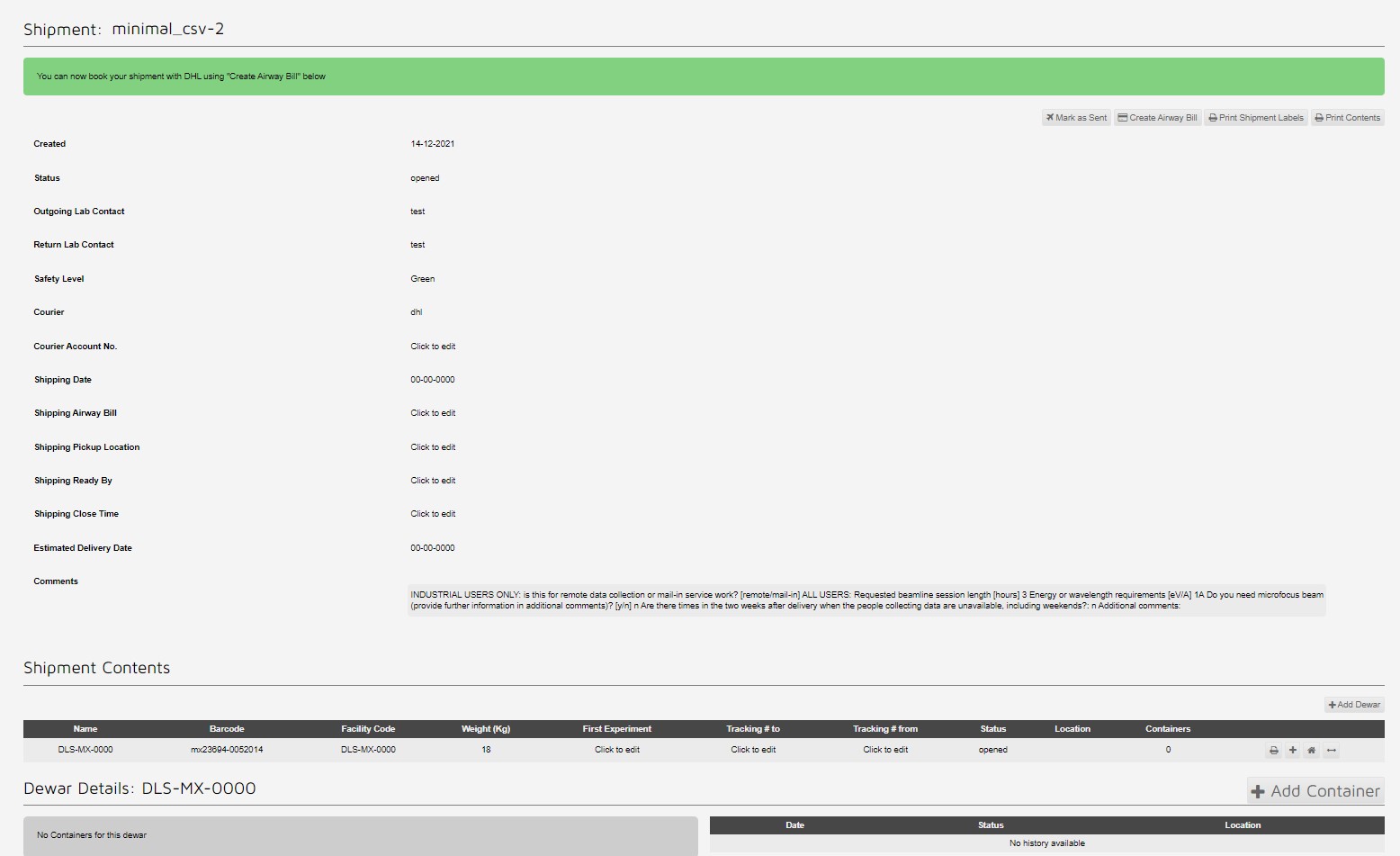
Then the upload script is run, similairly to below:
This creates a shipment with two pucks with 16 samples each:
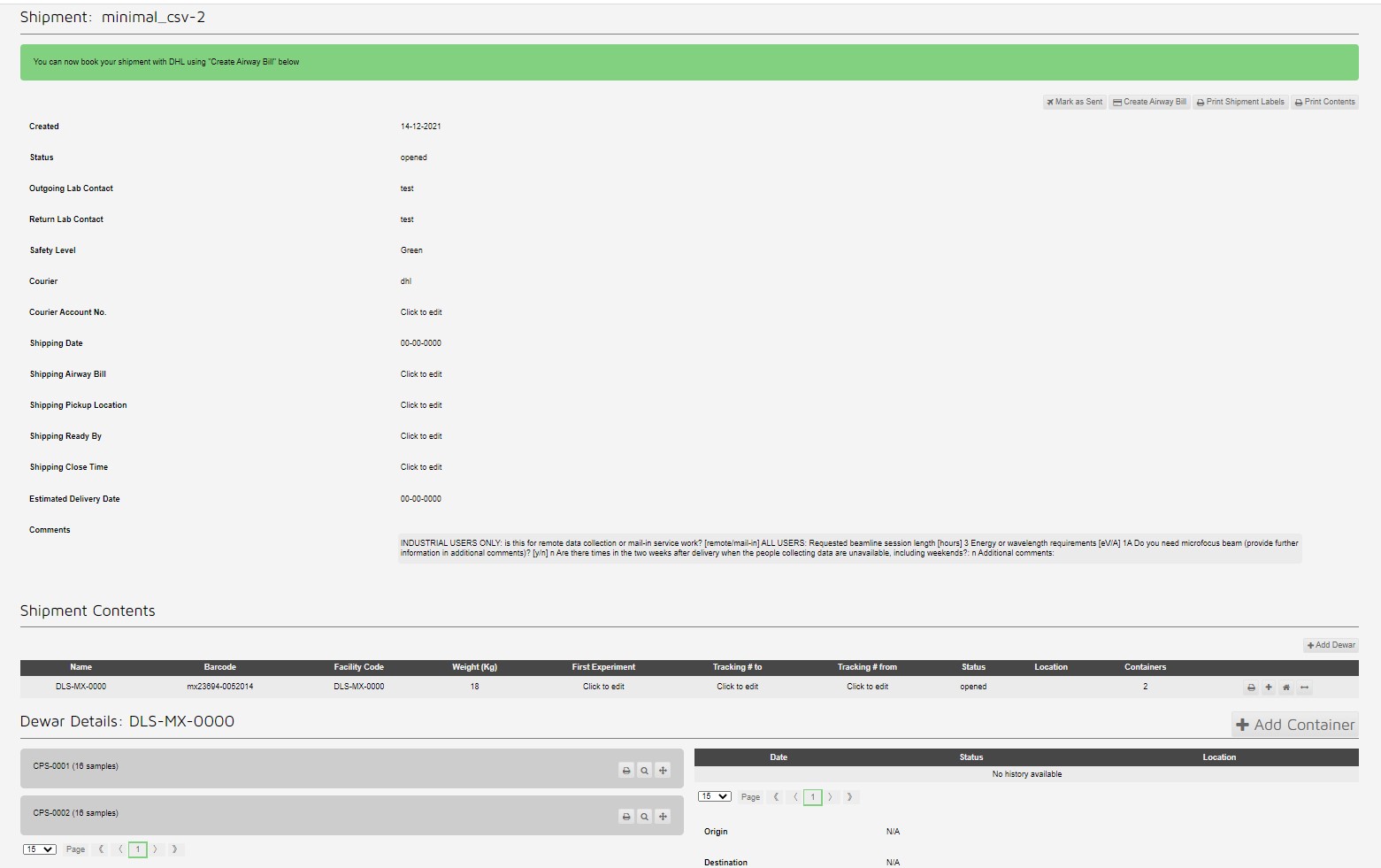
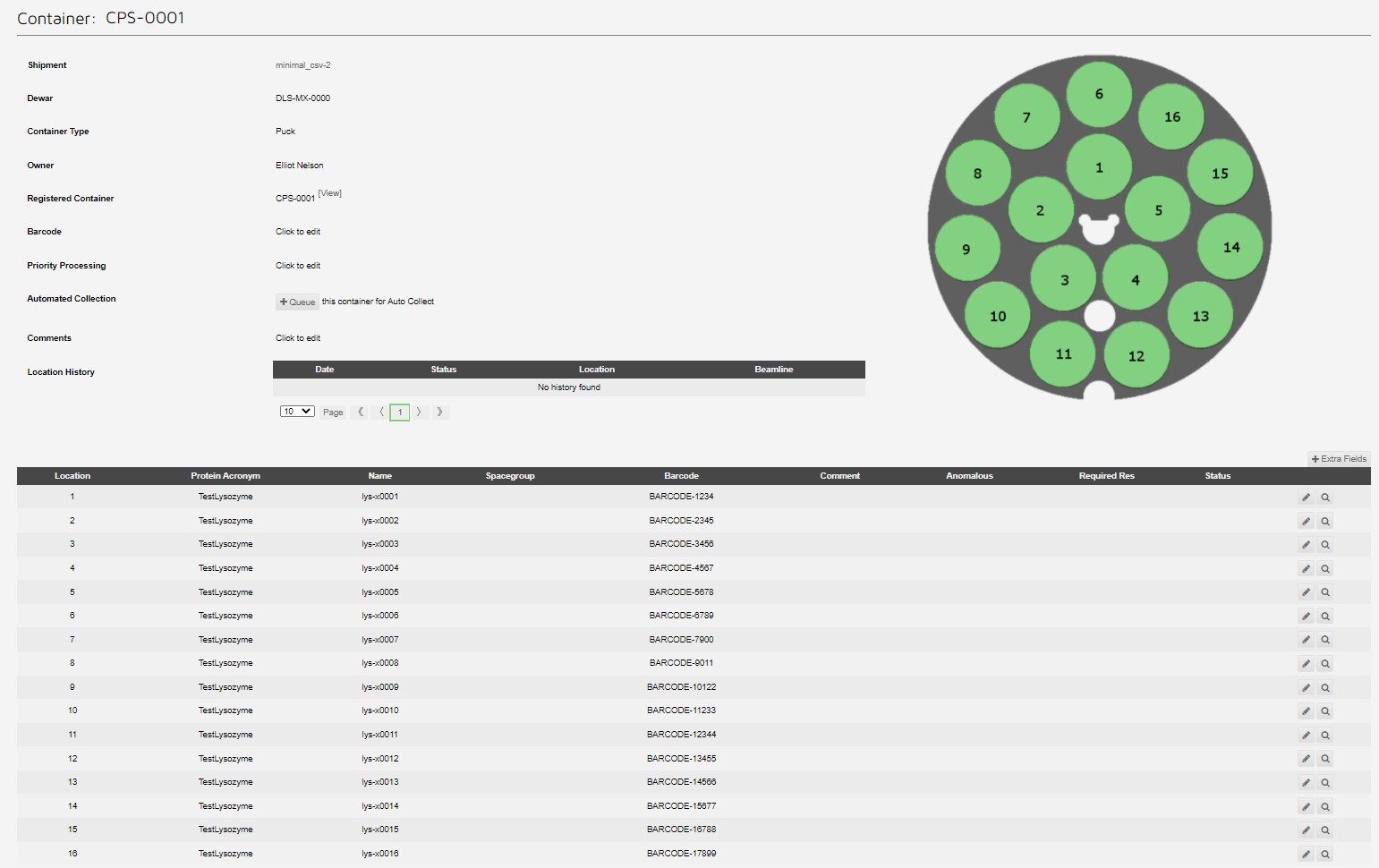
Which can be seen in the container view:
Unattended Data Collection example
Samples for unattended data collection (UDC) can be uploaded in avery similair manner, by specifying extra fields as in udc csv. The --UDC flag should be added when uploading, to queue samples for collection.
Giving a queued puck:
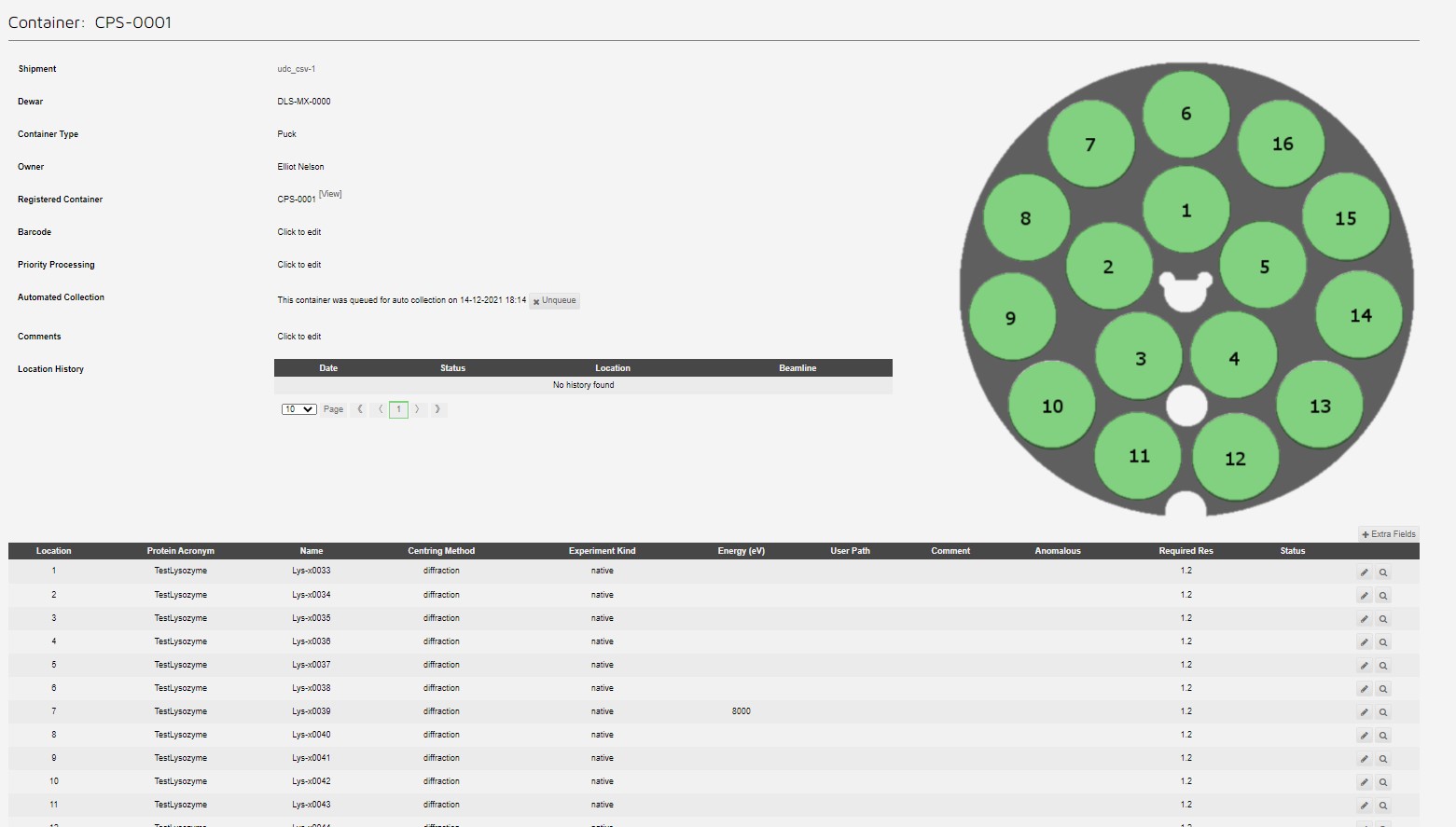
Screening strategy example
Template csv showing how to submit CSV with details of screening strategies, samples are set to match those defined in the below image:
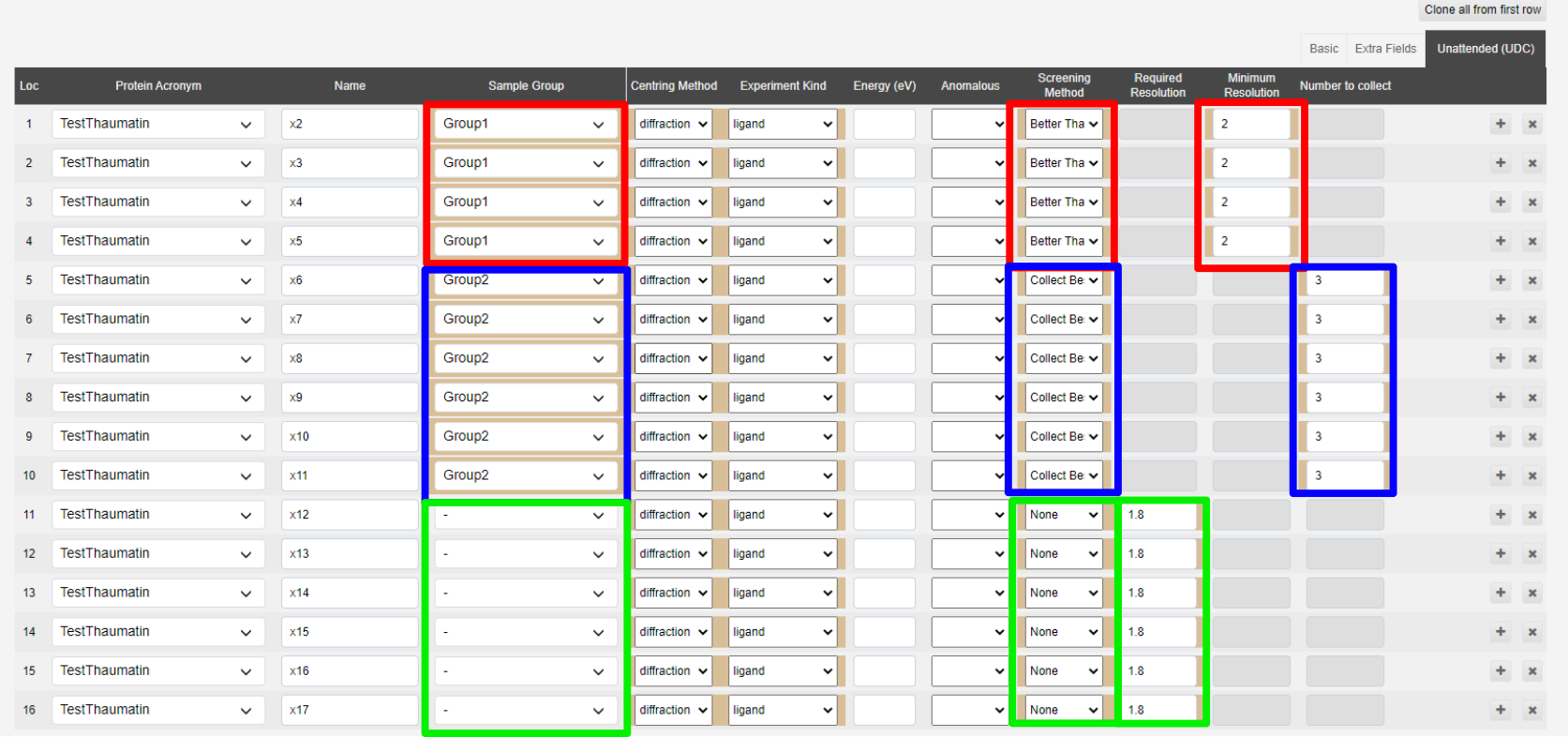
Error and warning messages
If not successful, the uploader will abort with an error message, or if there was a minor problem, then it will complete but with a warning message, such as:
- WARNING: Not setting lab contacts for shipment as the csv file owner <fedid> is not a lab contact for proposal mx23694.
Error Messages
- ERROR: The dewar code X is not a registered facility code for proposal Y
- ERROR: The container code X is not a registered container code for proposal Y
The dewar or puck code is not registered for the proposal, check it is correct in the CSV and contact [email protected] / [email protected] to register the dewar or puck.
- ERROR: Mandatory field %s not filled in. Required format is: %s
One of the mandatory fields (described above) is not filled.
- ERROR: One of these conditions must be met in order to upload the .csv file:
- The person uploading the .csv file (%s) must be a member of the proposal given inside the .csv (%s).
- The proposal given inside the .csv (%s) must be the proposal of the visit directory the .csv file is in (%s).
You must be a member of the proposal you are uploading samples to. Make sure you are added as investigator in UAS.
- ERROR: The proposal given inside the .csv (%s) does not exist in the database.
Please check that the proposal code and number is correct in the csv file
- ERROR: Mandatory field %s not filled in. (Only mandatory for first row.) Required format is: %s
One of the mandatory fields (described above) is not filled.
- ERROR: There are X occurrences of samples with name Y and protein acronym Z in this csv file
Check there are no duplicated samples.
- ERROR: The proteins must already exist in ISPyB - this one doesn't: acronym: %s
- ERROR: The proteins must have been approved - this one isn't: acronym: %s
The protein acronym to be used for sample upload to ISPyB must exactly match the protein acronym used in the ERA. Change acronym in the csv file or add sample ERA in UAS.
- ERROR: Sample %s in container %s is in an illegal location %s
- ERROR: Sample %s in container %s has an illegal non-integer location %s
- ERROR: Sample %s in container %s has location %s which is already taken.
- ERROR: Container %s has more than 16 samples
- ERROR: Sample with name %s already exists for protein with acronym %s in this proposal.
Please check your sample location in container (puck). Must be a unique value between 1-16. Check the sample is not duplicated.
- ERROR: Space group must be at least 2 characters long or be a positive integer: %s
- ERROR: Space group number must be in the range [1, 230]: %s
- ERROR: Space group %s not found in space group list.
- ERROR: If either of the unit cell parameters are defined, then all must be defined. Got %s for sample %s
- ERROR: All unit cell angles must be < 180 degrees. Got %s for sample %s
- ERROR: Unit cell volume must be positive. Got %s for sample %s with cell params %s
Please check that the space group and unit cell information conform to standards
- ERROR: Authorisation failure - the time delta is too large.
- ERROR: The userPath can be max 100 characters long, this one is longer: %s
For sample groups used with UDC, it is currently limited to uploading unique sample groups:
- ERROR: There is already a sample group for proposal mx23694 with name Group2
Upload script from other computers
An upload script can be used from bash environments (currently tested on ubuntu linux and bash shell installed on windows). This script uses the above upload script from diamond, but also automates the copying from the local computer to Diamond.
There may be issues with ssh/sftp configurations, if so please contact [email protected] / [email protected]. Note that the requirement to enter your password multiple times keeps the script more likely to work on differing configurations.


 Macromolecular Crystallography
Macromolecular Crystallography
