
David is particularly interested in improving MX beamlines with automation without compromising on quality of the research developed. This includes better ways to collect data with modern day pixel array detectors, strategies to collect complete datasets of diffraction data from multiple wedges on the same or different crystals, working with the challenges presented by small and ultra small X-ray beams (e.g. beam stability, small sphere of confusion, etc) as well as improvements on how we reduce our raw data from these different experimental setups.
Another area that David is keen is user experience at an MX beamlines from local at the synchrotron to remote at their labs. Improvements on how to design our experiments, on how to present the GUIs for data collection and how we organize the post experiment data archivers (raw and metadata) so that we can make a better use on the next experiments. The amount of data generated on a modern synchrotron due to advances on beam flux, size and detectors can be often overwhelming. These requires some thinking outside of the box and not present or do things the same way over and over.
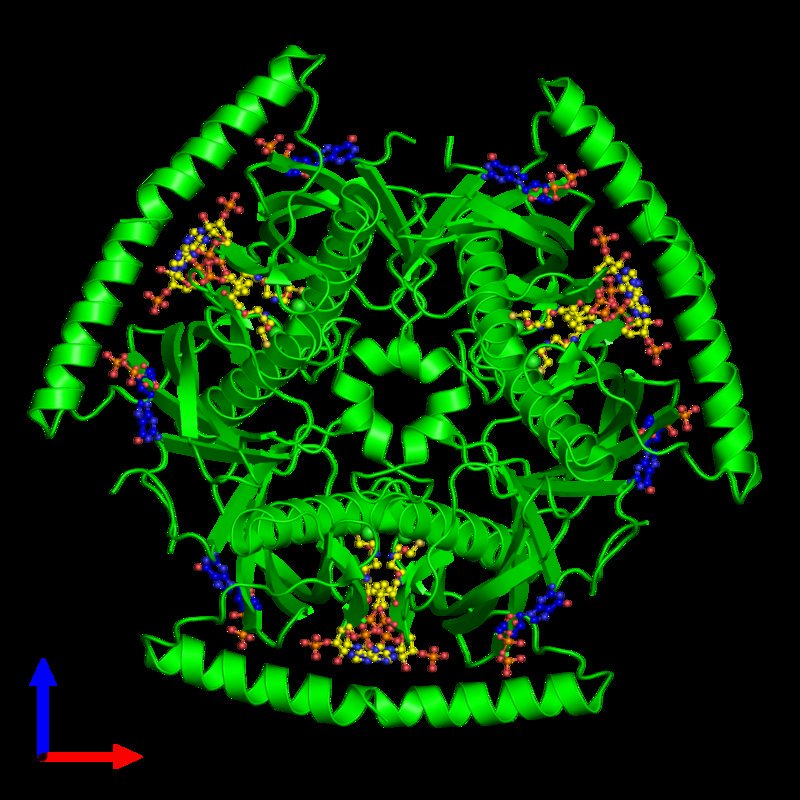
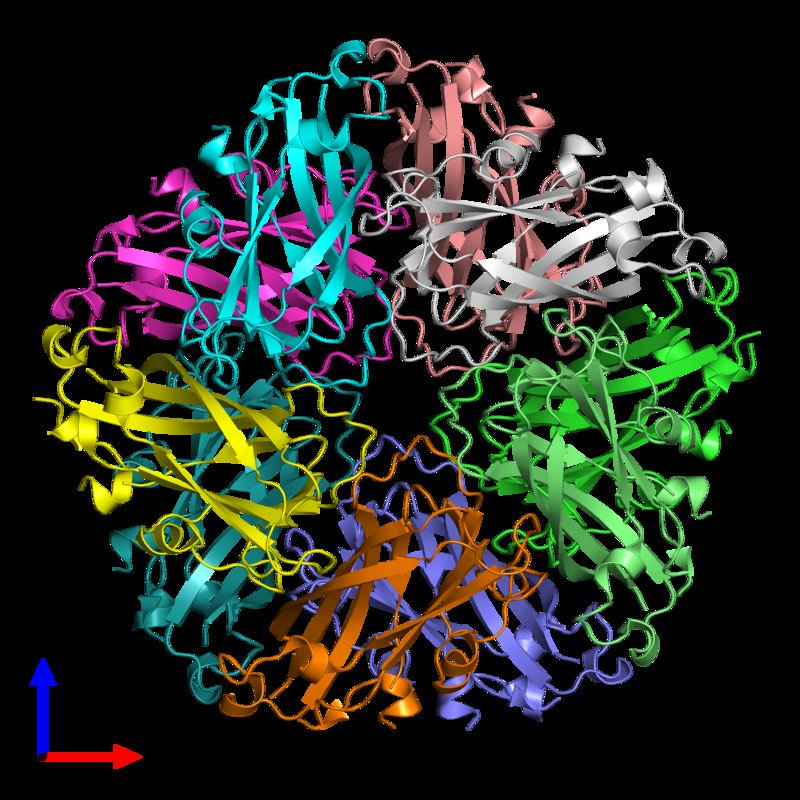
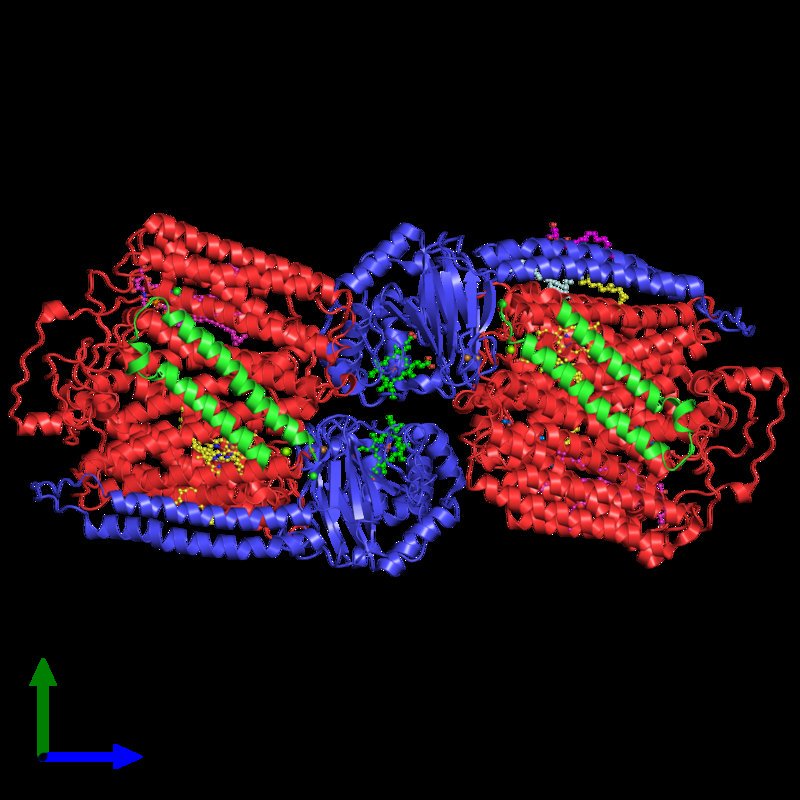
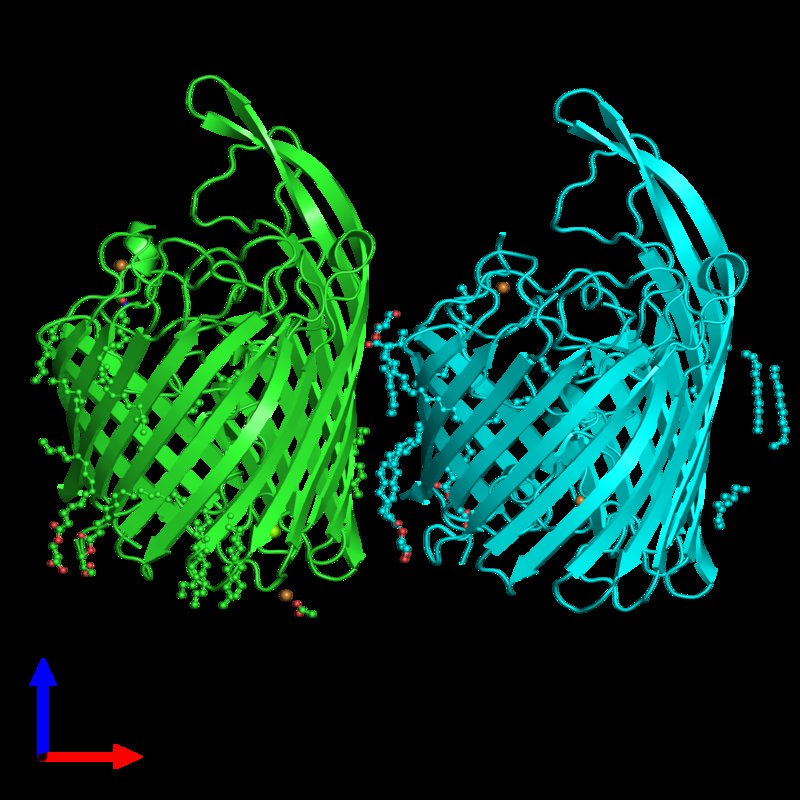
Finally, my structural biology research interests lie on the interfaces of cells and cell organelles: Understanding the biology how an important physiological agent is imported by a bacteria for survival, how energy is transduced to proton gradients across the biological membrane or how proton gradients are converted ATP, how viral capsids attach to cells or how viruses assemble inside their host cell, how cargo proteins import or export molecules into the nucleolus, mitochondria or Golgi apparatus is key to fundamental processes of medical research.



 Macromolecular Crystallography
Macromolecular Crystallography





