- Diamond Light Source
- News & Literature
- Print Publications
- Diamond News
- Spring 2016
- Focus on science
- > Transcriptional triumphs
Keep up to date with the latest research and developments from Diamond. Sign up for news on our scientific output, facility updates and plans for the future.
Groundbreaking work conducted at two of Diamond’s MX beamlines (I03, I04) has revealed the novel mechanisms employed by bacteria to regulate RNA production. For the first time, the structure and function of a bacterial transcription factor, known as sigma-54 (σ54), have been revealed by X-ray crystallography and new clues are now forthcoming as to how bacteria regulate their response to environmental stresses. These fascinating observations have recently been detailed in Science.
Gene transcription is a fundamental process essential for life and this process has been extensively studied in bacteria. Although much is known about the key player in transcription (the protein known as RNA polymerase), the transcription factors that interact with this protein to regulate the process are less well known. Of particular interest is the group of sigma (σ) factors, which are the primary controllers of RNA production in bacteria.
Of this group, one known as σ54 stands out as a unique target that warrants further investigation. All the genes under the control of σ54 are responsible for responding to stresses such as heat and osmotic shock, and the factor acts as an inhibitor to transcription. Professor Xiaodong Zhang of the study group explained: “σ54 is completely different to other sigma factors as it does not help to start transcription. In fact, it helps to dock RNA polymerase to DNA, but it inhibits transcription until additional activator proteins are recruited. This is a great system to study and we were keen to understand how this inhibition occurred.”
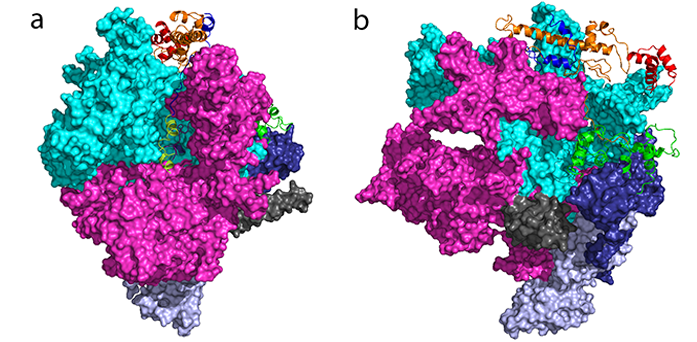
Crystal structure of RNA polymerase (depicted in surface view) in complex with σ54 (depicted in ribbon).
Scientists from Imperial College London, Peking University, University of Wisconsin, and Pennsylvania State University sought to study the intriguing σfactor in more detail using X-ray crystallography with the assistance of the dedicated staff at beamlines I03 and I04. The team crystallised the RNA polymerase-σ54 complex, but Prof Zhang admitted that the study ran into difficulties during X-ray crystallography: “It was quite a difficult project as the crystal diffracted poorly, but the new techniques at Diamond helped us to achieve the resolution we needed.”
The complex being studied was very large and fragile, so innovative methods developed at Diamond were employed to improve the diffraction patterns. Firstly, the sample’s crystalline properties were modified using the humidity control device, which helped to enhance the resolution. Secondly, due to the fragility of the crystal, a helical-rotation diffraction method was applied. Combining these techniques enabled the team to achieve a crystal structure at a resolution of 3.8 Å, which allowed them to effectively view the regions of σ54 that were interacting with RNA polymerase.
Prior to this study, the structure of σ54 was unknown, with no known structural homologues. This pioneering work has allowed this unusual transcription factor to be visualised for the first time. The study revealed that the structure was totally unique; consisting of three main domains that were connected by extended linkers. This knowledge in itself explains why the molecule could not be crystallised in isolation.
Furthermore, by studying σ54 bound to RNA polymerase, it could be visualised in situ. They discovered that σ54 interacted with huge areas of the polymerase using all of its three domains. A typical inhibitor of this kind might interact across one or two sites, but σ54 interacts at four key sites to prevent transcription. The researchers found that σ54 blocked the entry and exit points for DNA and it also blocked the region responsible for RNA synthesis and the exit point for the newly-made RNA.
The team are continuing to explore the mechanism of σ54, and have turned their attention to finding out how the inhibition is reversed through the action of activator proteins. Prof Zhang elaborated: “We know the region where the activator proteins work, but we want to see how the inhibition is released. We want to pinpoint the mechanism that allows σ54 to be disarmed.”
These remarkable discoveries have wide-reaching repercussions for the scientific community, both for the application of antibiotic development and for the development of potential control mechanisms for synthetic biology.
To find out more about using the I03 and I04 beamlines, or to discuss potential applications, please contact Principal Beamline Scientists Dr Katherine McAuley ([email protected]) or Dr Dave Hall ([email protected]) respectively.
Yang et al. Structures of the RNA polymerase-σ54 reveal new and conserved regulatory strategies. Science (2015). DOI:10.1126/science.aab1478
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.