Transcriptional repression in eukaryotes requires the recruitment of large protein complexes to gene promoters and enhancers. These complexes contain histone deacetylases, and other enzymes, that facilitate chromatin remodelling so as to repress gene transcription. SMRT and NCoR are homologous co-repressor proteins that are recruited to multiple repressive transcription factors. When isolated from HeLa cells, SMRT and NCoR purify as large complexes of between 1 and 2 megadaltons containing SMRT or NCoR, HDAC3, TBL1 and GPS2. The role of TBL1 and GPS2 in this complex have hitherto been poorly understood. We have used X-ray crystallography, NMR spectroscopy and a variety of biophysical and biological approaches to understand the assembly and function of the core SMRT/NCoR repression complex. These studies suggest that TBL1 plays a central role in mediating the assembly of the transcriptional repression machinery through acting as a structural scaffold to target HDAC3 activity to chromatin.
SMRT and NCoR are essential proteins both during development and for the maintenance of homeostasis. They are over 2,500 amino acids in length and are in the most part natively unstructured. They appear to serve as platform, or hub, proteins, that mediate transient interactions with a wide variety of repressive transcription factors. In addition to these transient interactions, SMRT and NCoR have a stable core complex containing HDAC3, a WD40 repeat containing protein, TBL1 (or its homologue TBLR1), as well as a protein called GPS2 (aka. AMF1). Using co-purification assays we precisely mapped the minimal regions of SMRT, GPS2 and TBL1 required for their three-way interaction. Although we were able to purify a stable ternary complex between the amino-terminal domain of TBL1 and the relevant interacting regions of SMRT and GPS2, we were not successful in obtaining diffraction quality crystals of this complex. However we obtained several crystal forms of the amino-terminal domain of TBL1 alone, which diffracted to between 2.2Å and 5Å. The structure resembles that of the LisH domain from Lis1. However, unlike Lis1, in all crystal forms TBL1 was observed to form a tetramer (Fig. 1) with a number of non-polar amino acids interdigitating at the interface between two TBL1 dimers. We subsequently confirmed, using size exclusion chromatography of wild-type protein and protein with a mutation at the tetramer interface, that TBL1 is also a tetramer in solution. Thus, it seems very likely that this is the physiologically relevant form of TBL1 in vivo.
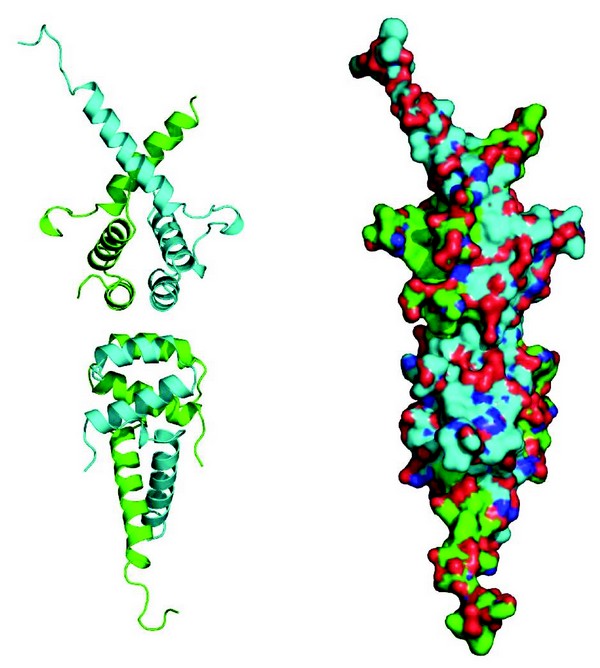
Figure 1: Cartoon and surface representations of the tetrameric oligomerisation domain of TBL1.
To understand the nature of the SMRT:GPS2 complex we determined the structure by NMR spectroscopy. These studies showed that the complex takes the form of a tight antiparallel coiled-coil (Fig. 2a). The antiparallel nature of the coiled-coil positions the TBL1 interaction regions in SMRT and GPS2 at the same end of the complex suitable for simultaneous interaction with TBL1. Through this means the two proteins cooperate to form a higher affinity complex with TBL1 than could be formed with either alone.
Examination of the TBL1 NTD structure revealed that each TBL1 NTD dimer has a pair of non-polar grooves on either side of the structure. This immediately suggested a potential mode of assembly for the complex (Fig. 2b). The importance of these non-polar grooves in mediating interactions with SMRT and GPS2 was confirmed by mutagenesis of residues at this surface. As expected these mutations abolished the interaction. Examination of the sequence of SMRT and GPS2 suggested that the proteins have in common a helical sequence motif: A x x ? H K/R x ? (where ? is a non-polar amino acid) which might mediate interaction with TBL1. This was confirmed for both proteins since mutations in the motif abolished recruitment to TBL1. Modeling and docking this motif (using HADDOCK software) from both SMRT and GPS2, with the TBL1 structure, revealed a unique mode of binding to the non-polar grooves of TBL1, with the key residues that define the interaction motif making important interactions at the interface (Figs. 2c and d).
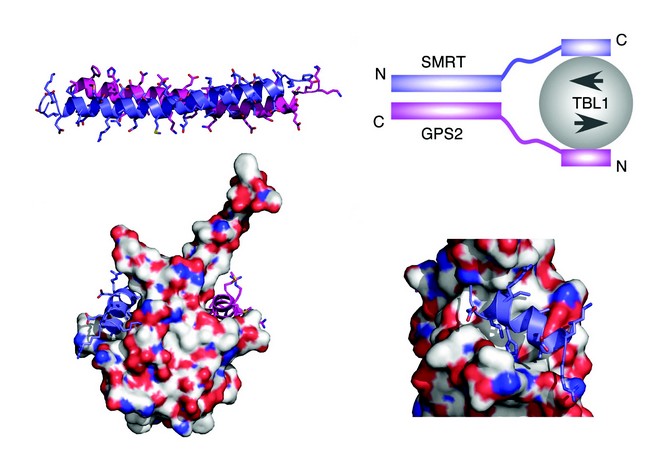
Figure 2: (a) NMR structure of the anti-parallel coiled-coil interaction between GPS2 (magenta) and SMRT (purple). (b) Schematic of the ternary SMRT:GPS2:TBL1 complex. (c) HADDOCK docking of the helical interaction domains of GPS2 and SMRT with the TBL1 dimer. (d) Detail of the SMRT peptide interaction with TBL1.
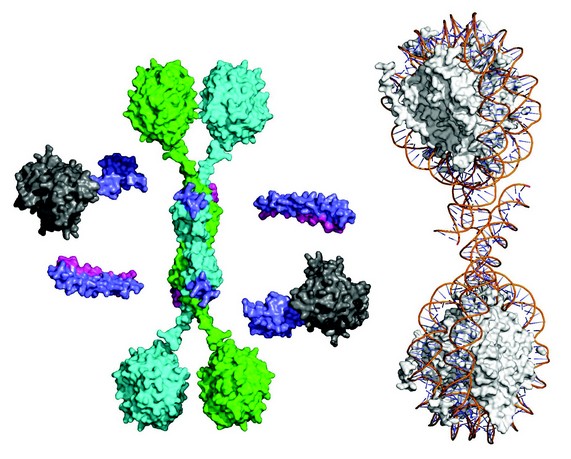
Figure 3: Synthesis of the structural data to yield a global illustrative model of the core SMRT complex including WD40 domains for TBL1 (green & cyan disks), HDAC (grey) and the SMRT-DAD (purple). A di-nucleosome is shown to the right to illustrate the relative scale of the chromatin substrate.
Taken together, our studies allow us to formulate an informative schematic of the core SMRT/NCoR repression complex (Fig. 3). In addition to the N-terminal tetramerisation domain TBL1 contains a C-terminal WD40 domain. It has been shown previously that WD40 domains are able to bind to histone tails and to intact nucleosomes. The tetrameric nature of the TBL1 suggests that the protein may serve as a scaffold for a multivalent chromatin-targeted repression machine that contains multiple corepressor and HDAC enzymes. Indeed, comparison of the anticipated size of the complex with a di-nucleosome also suggests that the complex may be able to simultaneously target HDAC3 activity to multiple nucleosomes.
Oberoi, J., Fairall, L., Watson, P., Yang, J-C., Czimmerer, Z., Kampmann, T., Goult, B., Greenwood, J.A., Gooch, J., Kallenberger, B.C., Nagy, L., Neuhaus, D. and Schwabe, J.W.R. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nature Structural and Molecular Biology. 18, 177-85, (2011)
Acknowledgements
The Wellcome Trust and UK Medical Research Council.
Beamlines used for data collection: I03 & I04 at Diamond Light Source and ID14.2 at the European Synchrotron Radiation Facility.