Plant diseases destroy ∼20 to 30% of annual crop production, significantly contributing to global food insecurity. Many plants are attacked by pathogens including viruses, bacteria, and fungi, resulting in disease. The study of the interaction between plants and pathogens allows researchers to better understand how plants protect themselves from pathogens, and to engineer disease resistant crops. A team of researchers from the John Innes Centre has used Diamond’s I03 beamline to investigate in atomic detail how a plant protein is specifically targeted by the blast fungus (Magnaporthe oryzae). Their results were recently published in PNAS. Their research is particularly important as this pathogen causes devastating losses in some of the most agriculturally important crops including rice, barley and wheat.
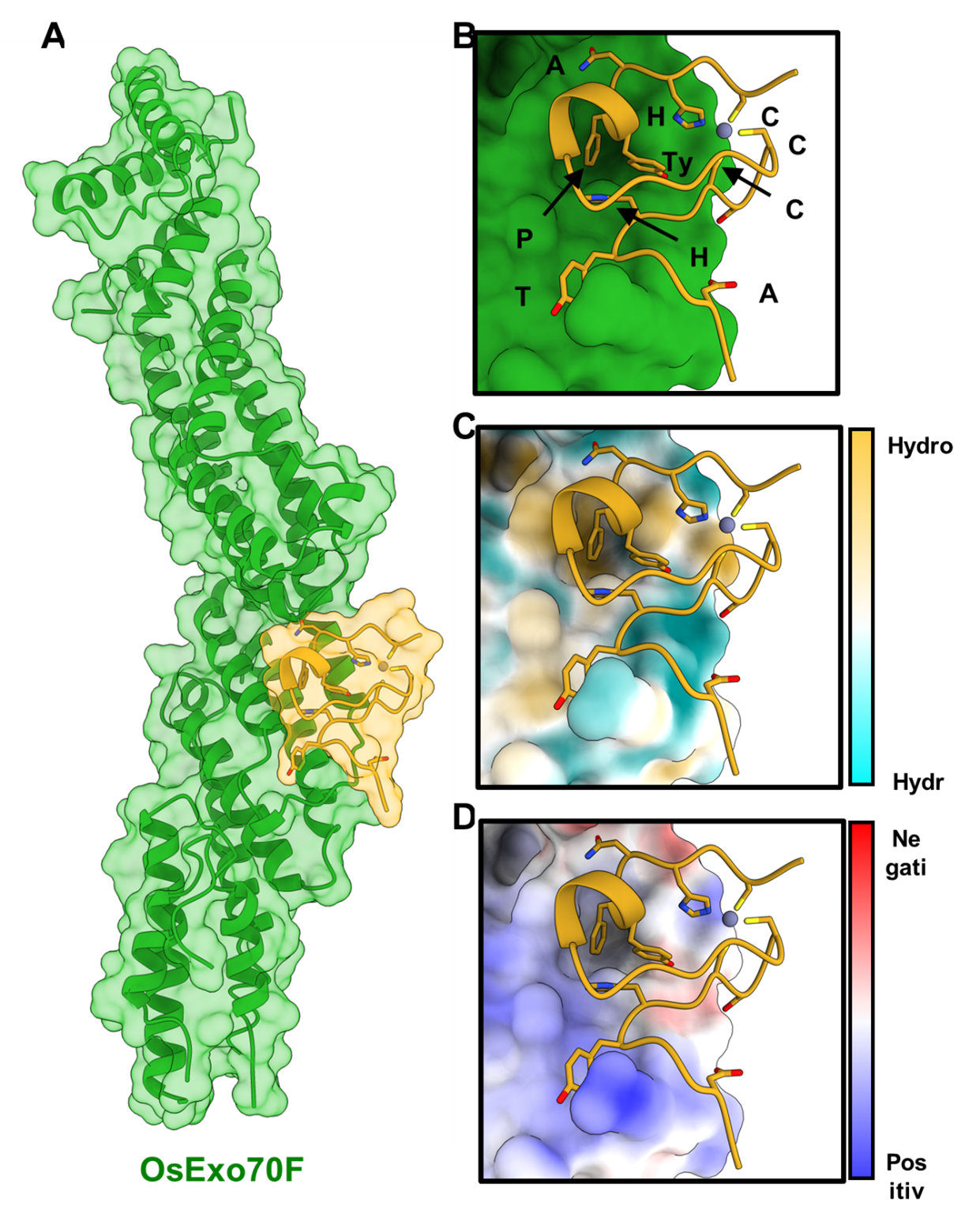
In all eukaryotes, there is a cellular pathway called exocytosis in which membrane-bound vesicles are delivered from intracellular compartments to the plasma membrane for the release of their contents into the extracellular space. This is essential for several key processes of the cell. Exocytosis is mediated by a protein complex called the exocyst. A protein from this complex, called Exo70, has expanded in plants, resulting in a diversity of protein sequence and multiple gene copies. This suggests that Exo70 proteins may have diversified and adopted specialised functions in plants. Specific plant Exo70 proteins have been associated with disease resistance to pathogens. One hypothesis is that certain Exo70 proteins are involved in the secretion of anti-pathogen compounds that would help the plant to resist the pathogen. The blast pathogen delivers effectors into the host to alter cellular processes, aiding successful colonisation. One such effector, AVR-Pii, interacts with two rice Exo70 alleles, OsExo70F2 and OsExo70F3, suggesting that the pathogen may target specific exocyst-mediated trafficking as a virulence-associated mechanism. Intriguingly, the interaction between AVR-Pii and Exo70 proteins is also an Achilles’ Heel for the pathogen as the complex is sensed by the plant immune system, triggering an immune response.
Mark Banfield is the leader of the Plant Health Programme at the John Innes Center. He says:
"To understand the complex interactions between pathogen proteins and their host targets, it is important to know their three-dimensional structures, including when bound to each other. This helps us understand function, and how this could be engineered for desirable outcomes, such as improved disease resistance. The Diamond Light Source is critical to our ability to derive X-ray diffraction data from protein crystals, ultimately allowing us to obtain a picture of protein structure."
To understand the structural basis of the interaction between Exo70 and AVR-Pii, the team of researchers, led by Prof. Banfield, used Diamond Light Source’s Macromolecular Crystallography Beamline I03. By doing so, they were able to determine with accuracy how the two proteins interact together. In addition, they reveal a zinc finger motif structure for the pathogen protein AVR-Pii, that has not been observed before. Based on the crystal structure derived from data collected at Diamond, the researchers performed specific mutagenesis of AVR-Pii to prevent interaction with Exo70 and correlated this with a loss of disease resistance in rice, confirming the importance of the interaction for the plant immune system to initiate a response. In the future, the researchers will target mutagenesis/engineering of plant Exo70 proteins, to further understand how recognition of pathogen proteins works, with an aim of extending the number of these proteins that can be detected by the plant immune system. They will also further investigate how the AVR-Pii/Exo70 interaction can act to promote disease when the plant immune system is unable to respond to this protein.
Juan Carlos De la Concepcion, previous PhD student in the Lab of Prof Banfield and first author on the study, commented:
Diamond Light Source has been a cornerstone of my research in the Lab of Prof. Banfield. Access to the beamlines allowed us to generate high quality data to determine macromolecular complexes between pathogen effectors and their targets in atomic detail. This allowed me to go one step further in answering biological questions regarding pathogenesis and plant immunity.
To find out more about the I03 beamline or discuss potential applications, please contact MX Group Leader Dave Hall: [email protected].
Video demonstrating the above interaction between the proteins. Credit Adam Bentham (JIC).
De la Concepcion J.C, et al. A blast fungus zinc-finger fold effector binds to a hydrophobic pocket in host Exo70 proteins to modulate immune recognition in rice. DOI: 10.1073/pnas.2210559119
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.