The DNA in human cells is regularly damaged, even by the body's normal cellular processes. For example, double-strand breaks (DSBs) in the chromosome can be caused by environmental exposure to radiation and chemicals, but may also result from cell metabolism. Left unrepaired or incorrectly repaired, DSBs can cause genomic rearrangements in the DNA, including deletions, translocations and fusions, that have the potential to cause cancer. Fortunately, the body has sophisticated pathways to repair damaged DNA. However, cancer cells can also use these repair pathways to survive radiation and chemotherapy treatments. In work recently published in Nature Structural & Molecular Biology, a team of scientists at Imperial College London and Washington University School of Medicine in St. Louis used high-resolution Cryo-Electron Microscopy (cryo-EM) structures to identify how a key protein kick-starts the DNA repair process.
This data could inform rational drug design to develop drugs that block DNA repair in cancer cells, making treatments more effective.
The last few decades have seen remarkable progress in our understanding and treatment of cancer. Although radiotherapy and chemotherapy can be very effective, some cancer cells can survive even high doses. Cancer cells can also make use of DNA pathways to repair themselves, but if we could develop drugs to disrupt those pathways, the cells would be more vulnerable to treatments.
Researchers from the Zhang Lab at Imperial College London study the signalling that happens on a molecular level when DNA is damaged. These signalling events are the early stages of the DNA repair pathway, alerting the cell to the need for repair. If a cell is in the process of replicating, the replication process must be halted until a repair is complete. In humans, ATR is a key protein involved in the start of the repair process.
Senior researcher Dr Luke Yates, explains:
We have a longstanding collaboration with researchers at the Washington University School of Medicine in St. Louis who work in yeast genetics. The Mec1 protein in yeast is essentially the same as human ATR, and so we can use it to study the DNA repair pathways.
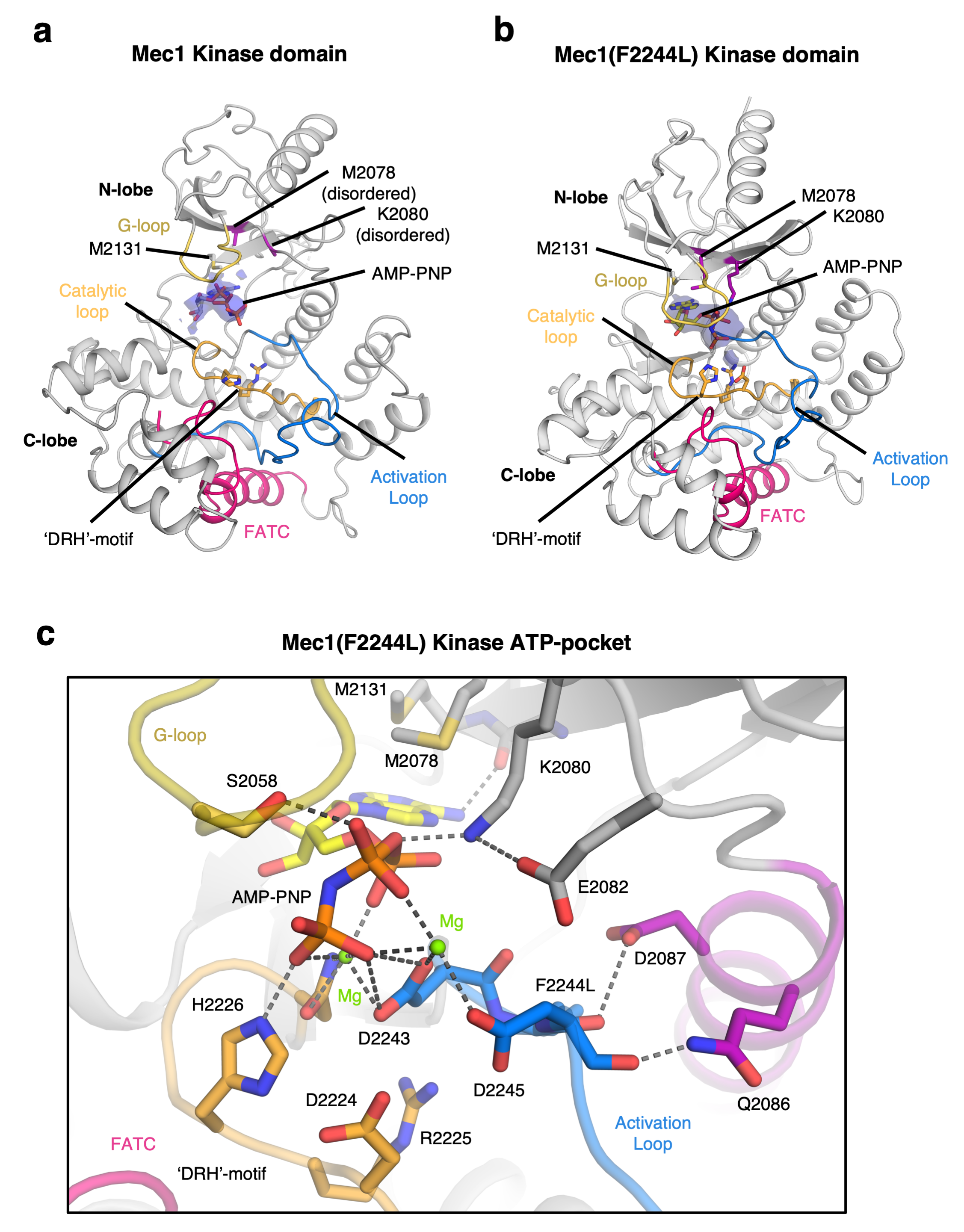
As this signalling molecule can halt cell replication, it is auto-inhibited (turned off by default). The Washington University team discovered a mutant version of Mec1 that is permanently on. We were, therefore, able to study the differences between the protein in its inactive and active states.
Film: Structural motion from comparisons between Mec1-Ddc2 and Mec1(F2244L)-Ddc2 to illustrate the global motion of the Mec1–Ddc2 complex upon activation. The video shows conformational changes across the Mec1–Ddc2 complex dimer interface when aligned on a single protomer. The trajectories between structures were calculated and visualized by morphing between AMP-PNP-bound wild-type auto-inhibited Mec1–Ddc2 and the AMP-PNP-bound constitutively active Mec1(F2244L)–Ddc2 mutant structure. Courtesy of Imperial College London.
The function of Mec1, and ATR, is to use a chemical called ATP to transfer a phosphate molecule from ATP onto other target proteins inside the cell’s nucleus. This ‘molecular labelling’ of specific proteins governing cell division signal to the cell to stop replicating its DNA and start repairing the damage. If we could ensure that the ATR protein remained inactive in cancer cells, then they wouldn't be able to begin the DNA repair process or stop replicating damaged DNA. This would make them more vulnerable to cancer therapies.
Clinical trials are underway for drugs, such as berzoserib, that target the ATP pocket of ATR to block its function. In recent UK trials these drugs can shrink advanced tumours in patients, and eliminate cancer in others, when used in combination with chemotherapy. However, these drugs were developed without knowledge of the protein's 3-dimensional structure. Understanding the structure of the protein, and how that changes when it becomes activated, could allow rational design of new and more potent drug candidates.
Dr Yates, added:
eBIC played a critical role in this research, as a national facility hosting a significant amount of expertise and cutting-edge equipment. It's fine-tuned to work exceptionally well. We collected around 20,000 micrographs for each version of the protein in just two allocated shifts at eBIC, yielding high-resolution cryo-EM reconstructions.
Back at Imperial, Dr Yates was able to process the micrographs to generate a 3-dimensional reconstruction of the protein structures. These structures offer new insights into how the protein is switched on and off. Complemented by genetics and biochemical analysis conducted by collaborators in Washington School of Medicine in St. Louis, and guided by structural insights, the team were able to identify which parts of the protein were important for its job inside the cell. Together these data will be invaluable for drug development.
Prof. Zhang, said:
Now we know what the activated state looks like and key changes that are required for the protein to turn on, the next steps for this research are to investigate how Mec1 is recruited to the damaged site, how different activators induce these changes, and how Mec1 selects and acts on its substrates.
To find out more about cryo-EM, or to discuss potential applications, please contact Alistair Siebert: [email protected].
Tannous EA et al. Mechanism of auto-inhibition and activation of Mec1-ATR checkpoint kinase. Nature Structural & Molecular Biology. Nov. 9, 2020. DOI 10.1038/s41594-020-00522-0.
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.