Keep up to date with the latest research and developments from Diamond. Sign up for news on our scientific output, facility updates and plans for the future.
Related publication: Zoll S., Lane-Serff H., Mehmood S., Schneider J., Robinson C. V, Carrington M. & Higgins M. K. The structure of serum resistance-associated protein and its implications for human African trypanosomiasis. Nat. Microbiol. 3, 295–301 (2018). DOI: 10.1038/s41564-017-0085-3
Publication keywords: African trypanosomes; Innate immunity; Apolipoprotein L1; SRA
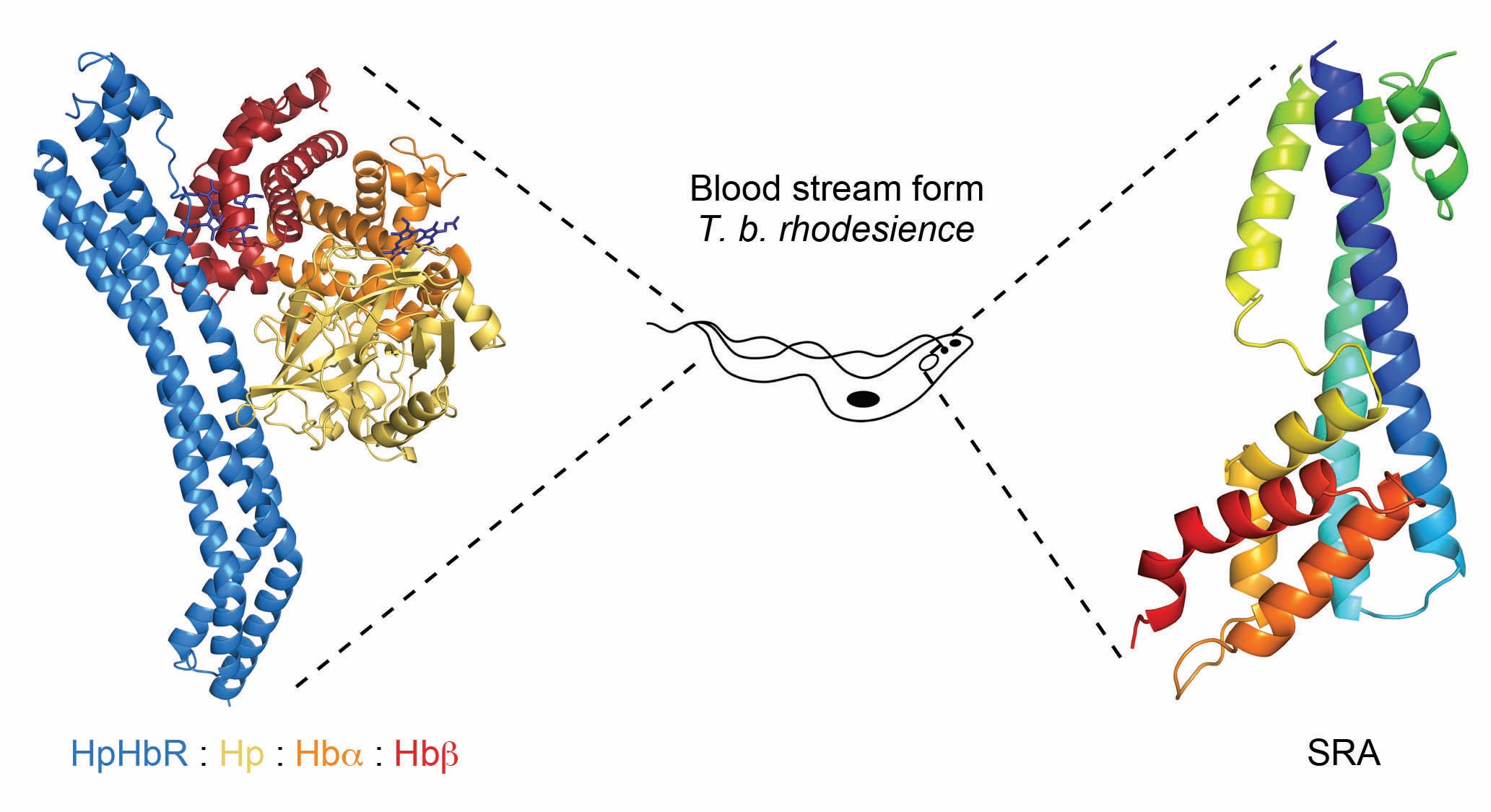
African trypanosomes are single-celled eukaryotic pathogens that are transmitted by tsetse flies, and live within the blood streams and tissue spaces of infected mammals. Infection causes disease, including the debilitating wasting illness of cattle, nagana, which restricts the productivity of livestock in sub-Saharan Africa. While there are numerous species of African trypanosome, only two subspecies, T. b. gambiense and T. b. rhodesiense, can proliferate in humans. This is due to lytic factors, present only in humans and some other primates, which are taken up by trypanosomes and causes their death1. Human-infective species are able to negate the action of this lytic factor. This confers the ability to grow within infected humans, and to cause the disease Human African trypanosomiasis, also known as sleeping sickness. The aim of our work is to understand how human-infective trypanosomes inactivate the lytic factor, and then to determine whether we can make them susceptible to lytic factor mediated killing.
Our first contribution towards these goals used beamline facilities at Diamond Light Source to understand how lytic factor is taken up by trypanosomes. The lytic factor is a complex of several components, including the pore-forming protein, apolipoprotein LI (ApoLI), which acts as the toxin and a complex of haptoglobin related-protein bound to haemoglobin (HprHb). Trypanosomes have a receptor on their cell surface that binds to haptoglobin-haemoglobin (HpHb), allowing them to scavenge this valuable nutrient from the blood. However, the presence of HprHb in lytic factor allows this toxin to hitch a ride on the receptor, taking it into the trypanosome cell, where it mediates its toxic effect. We used beamlines I03 and I04-1 to solve structures of HpHb receptors from human-infective and non-infective trypanosomes, alone and also bound to HpHb2-4. This revealed how the lytic factor is recognised by trypanosomes. It also helped us to understand how T. b. gambiense becomes human-infective, as it has a mutation of the HpHb receptor which reduces lytic factor uptake. Our structures revealed how this mutation decreases the affinity of the receptor for lytic factor, and how it helps to protect this trypanosome subspecies from destruction2,3.
References:
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.