Keep up to date with the latest research and developments from Diamond. Sign up for news on our scientific output, facility updates and plans for the future.
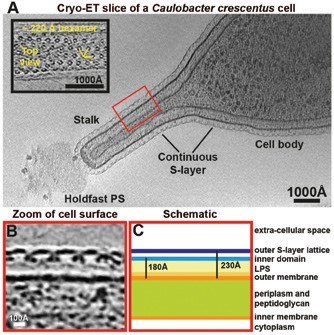
S-layer proteins (SLPs) are a diverse class of molecules found on many prokaryotes including Gram-positive and Gram-negative bacteria and the majority of archaea1. SLPs assemble to form planar sheets called S-layers on the surface of cells, where they are anchored, usually through non-covalent interactions with other surface molecules such as lipopolysaccharide 2. The propensity of SLPs to form sheets in solution and their absence from early model organisms such as Escherichia coli have hampered high-resolution structural biological analysis of this abundant and important class of molecules3. We turned to Caulobacter crescentus, a well-studied Gram-negative alphaproteobacterium with a characteristic ultrastructure. The cells are completely covered with an S-layer that is continuous between the cell body and the stalk (Fig. 1a). Cryo-ET imaging showed that the density corresponding to the S-layer of C. crescentus CB15 is almost perfectly hexameric (Fig 1a, inset) with a ~220 Å repeat distance. Two discrete densities were observed in the S-layer, the outer highly connected S-layer lattice and the discrete inner domains located around the centres of the hexamers (Fig. 1b-c).
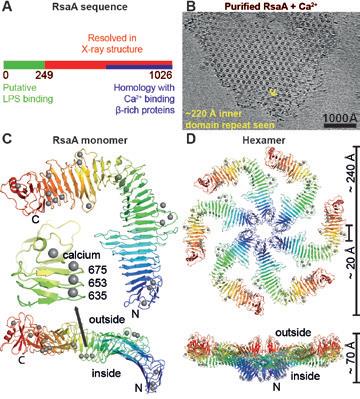
All residues in the RsaA X-ray structure were well ordered, reflecting a requirement that the S-layer lattice and the RsaA protein must avoid flexible loops, as they would be easy targets for extra-cellular proteases (Fig 2c). Amino acid side chains buried inside the β-helix domain of RsaA are either hydrophobic, or are negatively charged and co-ordinated tightly to structural Ca2+ ions (Fig. 2c-d, grey spheres), with very few exceptions. Some Ca2+ ions are in close proximity to the central pore of the hexamer (Fig. 2d), where they appear to stabilise the hexameric interface.
To relate the RsaA X-ray structure to actual S-layers found on the surface of cells, the structure of the C. crescentus S-layer using electron cryotomography (cryo-ET) of cells was resolved and subsequent sub-tomogram averaging also. The final refined 7.4 Å structure allowed us to locate different domains of the S-layer in the map and identify secondary structure elements (Fig. 3a). The outer S-layer lattice is highly inter-connected and consists mainly of β-sheets while the inner domain is discrete.
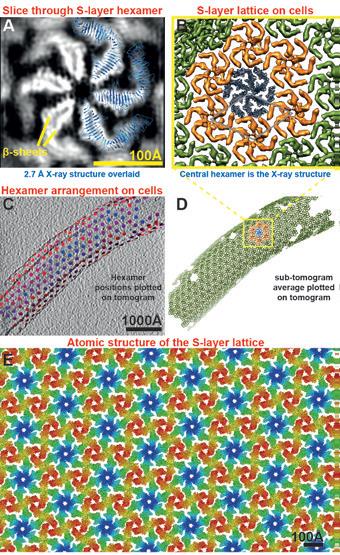
We then docked the hexamer from the RsaA249-1026 X-ray structure into the cryo-ET density map of the outer S-layer lattice as a rigid body. The docked atomic model shows that the lattices formed in the crystals through crystal packing and on cells are extremely similar and allows us to describe the arrangement of the S-layer lattice on cells at the atomic level (Fig. 3b-e). In the X-ray lattice, the hexameric interface contains amino acids bound to Ca2+ ions. We propose that Ca2+ ions bound near this interface are essential for the assembly of the RsaA hexamer. There is a strong dimeric interface and a less extensive trimeric contact is also observed in the lattice. Ca2+ ions are located near both the dimeric and trimeric interfaces in the lattice, possibly increasing the Ca2+ dependency of S-layer assembly.
Related publication: Bharat TAM, Kureisaite-Ciziene D, Hardy GG, Yu EW, Devant JM, Hagen WJH, Brun YV, Briggs JAG, Löwe J. Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nature Microbiology 2, 17059, doi:10.1038/nmicrobiol.2017.59 (2017).
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.