Related publication:
Hengrung N, El Omari K, Serna Martin I, Vreede FT, Cusack S, Rambo RP, Vonrhein C, Bricogne G, Stuart D, Grimes JM, Fodor E. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 5, 114-117, doi:10.1038/nature15525 (2015).
Keywords:
Influenza virus; Viral RNA polymerase; Replication.
Influenza viruses are a major public health issue, with seasonal viruses responsible for approximately half a million deaths worldwide each year and new pandemic viruses constantly threatening to emerge. Although well-known pathogens, the way that influenza viruses replicate is not yet understood at a molecular level. The viruses copy their RNA genome using a viral RNA polymerase, therefore understanding the structure of this polymerase is fundamental to understanding the mechanism of viral replication.
Crystals of the polymerase were examined at Diamond Light Source using the Macromolecular Crystallography (MX) beamlines, in particular I03, I04 and I24, to solve the structure, and the High Throughput SAXS beamline (B21) to monitor the global conformational changes. The Small-Angle X-ray Scattering (SAXS) data revealed the structure of the molecule to be highly sensitive to its environment. The RNA polymerase of the influenza virus was shown to be a highly flexible and dynamic molecule that could change its structure in response to subtle changes in its environment. This finding is not surprising when considering the complex chemistry that the polymerase performs during the viral lifecycle. Further careful work will be required to work out what additional conformations are accessible under what conditions to build an extensive mechanistic picture of the polymerase. Once complete, these studies may facilitate the development of antiviral compounds that target the polymerase.
Soft Condensed Matter Village & MX Village | Beamlines B21/I03/I04/I24
Influenza viruses are a major public health issue; with seasonal viruses responsible for approximately half a million deaths worldwide each year, and new pandemic viruses constantly threatening to emerge. This makes developing a molecular understanding of the workings of the virus a pressing issue. Virus particles are essentially machines that infect and replicate within cells. This involves making new copies of viral genome, which for influenza is single-stranded ribonucleic acid (RNA), and transcribing this genomic RNA to direct the production of the viral protein components. Like other similar viruses, influenza carries its own polymerase (FluPol), which catalyses both of these processes. As such, FluPol lies at the heart of the influenza virus lifecycle and has been the focus of a prolonged research effort to understand its structure and mechanisms. Ultimately, this may facilitate the development of antiviral compounds that target the polymerase.
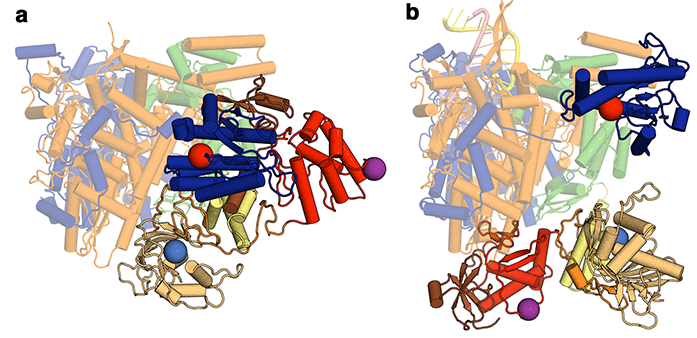
Figure 1: The structural differences between (a) apo and (b) RNA promotor bound influenza polymerase. The invariant portion of the molecule is pale coloured, whilst the domains that move are drawn in brighter colours.
This paper describes an X-ray crystal structure of the influenza C virus polymerase, complementing work from Stephen Cusack’s lab who last year released structures of the influenza A and B virus polymerases co-crystallised with viral promotor RNA1,2. Together, these have provided the first molecular views of the complete FluPol. However, the structures are not all equivalent. There are striking differences between the model described in this paper, which was obtained in the absence of viral RNA, and the previously deposited polymerase-RNA complexes. Though the fold of each of the domains within the 255 kDa heterotrimeric complex of FluPol is similar, the arrangement of these domains is dramatically altered between the unbound (apo) form and RNA-bound complexes (Fig. 1). More specifically, these rearrangements occur within the C-terminal two thirds of one subunit and the N-terminal quarter of another, with the effect that a binding pocket required for transcriptional activity is blocked in the apo conformation. FluPol must carefully balance the two quite different and intricate tasks of replication and transcription, so it has been long suspected that conformational changes are critical to its function. However the scale of the conformational changes involved, as revealed by this work, was still not anticipated.
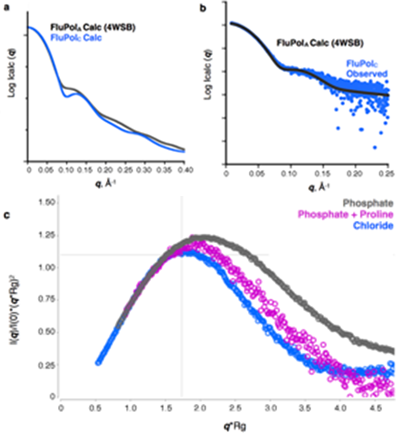
Figure 2: SAXS analysis of influenza C polymerase.
In order to further explore the functional significance of these conformational changes, B21 was used to probe the solution-state structure of FluPol. The theoretical X-ray scattering profiles of the apo and RNA-bound FluPol complex are distinct (Fig. 2a), meaning that these forms should be readily distinguishable. The onsite size-exclusion chromatography setup at the beamline also ensured high sample quality, which was particularly important given the tendency of the polymerase to form aggregates over time.
The scattering profile of the apo influenza C polymerase was measured and compared to profiles calculated from the crystal structures. Surprisingly, the measured profile closely corresponded to that of an RNA-bound FluPol (Fig. 2b), indicating that the majority of polymerase in solution resembled this, instead of the apo form. This has several implications. Firstly, it confirms that the alternative conformation observed in the crystal was not simply an influenza virus type difference, making it likely that A and B polymerases can also adopt similar conformations. Thus the conclusions are relevant across all virus strains. Secondly, it indicates that the polymerase must be highly flexible, since it can convert between at least two radically different conformations in the absence of RNA. This is plausible because the major differences between apo and RNA-bound forms are all in regions distal to the RNA-binding site. Measurements of the polymerase were also taken in different buffers and this too affected its conformation, further indicating that the energy barrier between different conformations is low. These results suggest that FluPol may be poised between several different conformations, with only subtle environmental changes needed for a particular conformation to be favoured.
In context, the SAXS data revealed that the regulation of FluPol is complicated. It is known that certain viral RNA sequences are required to make FluPol competent for transcription, and the crystal structure seemingly explains why this is the case; these RNAs cause a conformational shift from an inactive form to an active one. However, during the SAXS experiments without RNA, the majority of FluPol resembled the active form. This incongruity could be explained by the salt concentration used during the SAXS experiments, which was higher than that present in the crystallisation drops, possibly influencing the conformation. It is also worth noting that subsequently published work, again involving crystallography and SAXS, with a different FluPol has largely corroborated many of the observations made here. Though many questions about FluPol still remain unanswered, what is clear is that it is an extremely flexible protein complex and that this flexibility is fundamental to its function. Further careful work will be required to work out what additional conformations are accessible under what conditions to build a complete mechanistic picture of the polymerase. Continued access to B21 will be important, as its clear that SAXS will be an invaluable technique in this future effort.References:
- Hengrung, N. et al. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 527, 114–117, doi:10.1038/nature15525 (2015).
- Pflug, A., Guilligay, D., Reich, S. & Cusack, S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516, 355–360, doi:10.1038/nature14008 (2014).
Funding acknowledgements:
This work was supported by Medical Research Council (MRC) grants MR/K000241/1 (to E.F.), G1100138 (to F.T.V.) and G1000099 (to D.I.S.), Wellcome Trust Studentship 092931/Z/10/Z (to N.H.), an MRC Studentship (to I.S.M.), and Wellcome Trust administrative support grant 075491/Z/04.
Corresponding author:
Professor Jonathan Grimes, University of Oxford, jonathan@strubi.ox.ac.uk.


 Diamond Annual Review 2016
Diamond Annual Review 2016

