 | ||
Structure of an antibacterial peptide ABC transporter |
Beamline I02 Scientific Highlight
In order to better understand the detailed mechanism of this bacterial immunity process and also understand how bacteria confer resistance to peptide-based antibiotics, it was important to understand the structure of different classes of ABC exporters. Studies were conducted on the ABC exporter McjD across all the Macromolecular Crystallography (MX) beamlines, with the final structure being determined on I02. McjD can be used as a model protein to study antibacterial peptide exporters.
Microcins are gene-encoded antibacterial peptides of low molecular weight <10 kDa, produced by Enterobactericeae. They are secreted under conditions of nutrient exhaustion and exert potent antibacterial activity against closely related species1 which provides them with more nutrients for survival. The microcin MccJ25 is a plasmid-encoded, ribosomally synthesised antibacterial peptide consisting of 21 amino acids. Four genes are required for the biosynthesis of MccJ25, mcjABCD; the mcjA encodes the linear 58 amino acid precursor of MccJ25 that is ribosomally synthesised, mcjB and mcjC encode proteins involved in the post-translational modification of McjA to the lasso structure. The mcjD encodes for an ABC transporter that is required for both MccJ25 secretion and self-immunity towards MccJ25. Since the genes are under the same operon, production and export from the cell is very efficient and prevents toxic levels of MccJ25 from building up. The tertiary structure of MccJ25 has been elucidated and shows a very unique lasso structure (Fig. 1a). Inside the cell, MccJ25 inhibits the bacterial RNA polymerase.
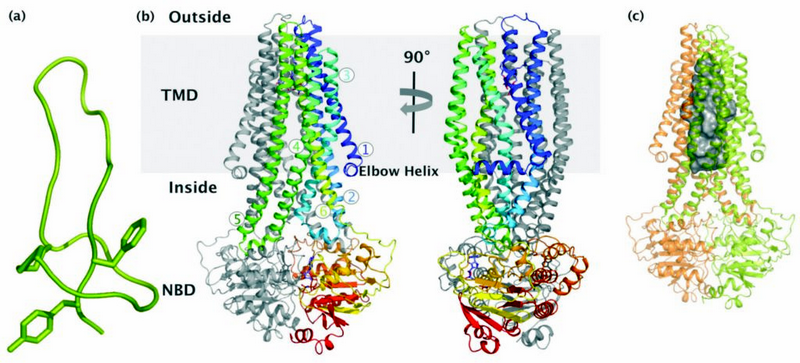
Figure 1: Crystal structure of the antibacterial peptide ABC exporter McjD; (a) NMR structure of the antibacterial lasso peptide MccJ25. (b) McjD adopts a nucleotide bound outward occluded kbeis@imperial.ac.uk conformation. The structure is viewed in the plane of the membrane. The transmembrane helices of one subunit are coloured from blu
McjD structure
We have determined the crystal structure of McjD from Escherichia coli at 2.7 Å resolution2 in complex with the ATP analogue 5′-(β,γ-imido)triphosphate (AMP-PNP) (Fig. 1b). McjD is a homodimer and contains an N-terminal transmembrane domain (TMD, 6 TM helices) and a C-terminal nucleotide binding domain (NBD). The dimer interface at the TMDs is formed between TM2 and TM5/TM6 from one monomer, with the equivalent TM5/TM6 and TM2 from the opposite monomer that form a large cavity with a volume of ∼5,900 Å3 (Fig. 1c). This cavity can accommodate one molecule of MccJ25. Two detergent molecules (nonyl-glucopyranoside) that probably stabilise the transporter are also found inside the cavity.
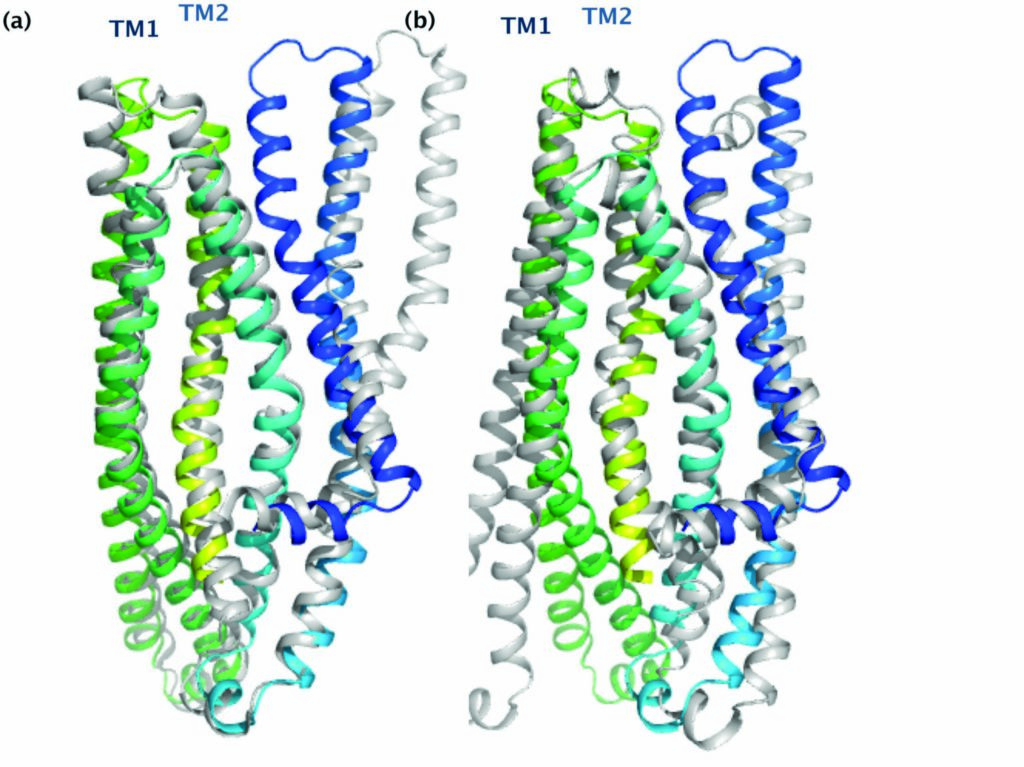
Figure 2: Comparison of McjD with other ABC exporters. The TMs of McjD can be superpositioned to (a) Sav1866 (outward open, grey) and (b) Vibrio cholerae MsbA (inward closed apo, grey) with an rmsd of 2.3 Å and 2.4 Å respectively. McjD shows structural similarities to the cytoplasmic TM region of Sav1866 and the periplasmic region of MsbA. TMs 1 and 2 show the most significant conformational changes between McjD and Sav1866.
Comparison with other ABC exporters
Comparison of the McjD TMD with the outward-open structure of Sav18663 and inward-closed structure of MsbA4 (Fig. 2) revealed that the transporter is occluded at both the cytoplasmic and periplasmic sides. The Sav1866 and MsbA structures form intertwined dimers between TMs1 and 2 of one subunit and TMs 3-6 of the other subunit. In Mcjd, no subunit intertwining is observed; TMs 1 and 2 have rotated by 26° and shifted by 6 Å at extracellular loop 1 relative to Sav1866. TM1 is closer to TM6 of the same monomer and TM1 of the adjacent monomer. TM2 is close to TM5 from the adjacent monomer. The conformation of the TMs occludes the periplasmic side. McjD has three salt bridges between Lys80 and Glu301 (from the opposite monomer), Arg141 and Glu309, and Arg195 and Asp135 that appear to stabilise the interaction of TM2 to TM6 (of the opposite monomer), TM3 to TM6, and TM3 to TM4 in our occluded state. The outward-open Sav1866 has salt bridges between Glu78 from TM2 and Arg295 from TM6 of the opposite monomer.
Mechanistic implications
ABC exporters alternate between inward- and outward-facing conformations in order to export their substrates driven by the hydrolysis of ATP (Fig. 3). Until recently, the proposed mechanism was an alternating access model in which TMs 4 and 5 are exchanged between the TMDs of the half-transporters in the inward-facing dimer (state i and ii), whereas the TMs 1 and 2 are exchanged in the outward-facing dimer (state iv). Substrate binds to the inward-open facing conformation and in turn it triggers binding of ATP (state i and ii). However, the previous models did not explain in detail how the transition between the two conformations occurs. Since the McjD structure has similarities to the closed cytoplasmic side of outward-open Sav1866 and MsbA, and the closed periplasmic side of inward-open apo MsbA, it represents an intermediate state between the two conformations. After the ligand has left the outward-open cavity, TMs 1 and 2 rotate away from TMs 3–6 of the opposite monomer to form the outward-occluded state without TMD intertwining (state v). The transporter ‘resets’ by adopting the inward-open conformation after ATP hydrolysis that disrupts the NBD dimer interface and induces intertwining of TMs 1–3, 6 from one monomer with TMs 4 and 5 from the opposite monomer.
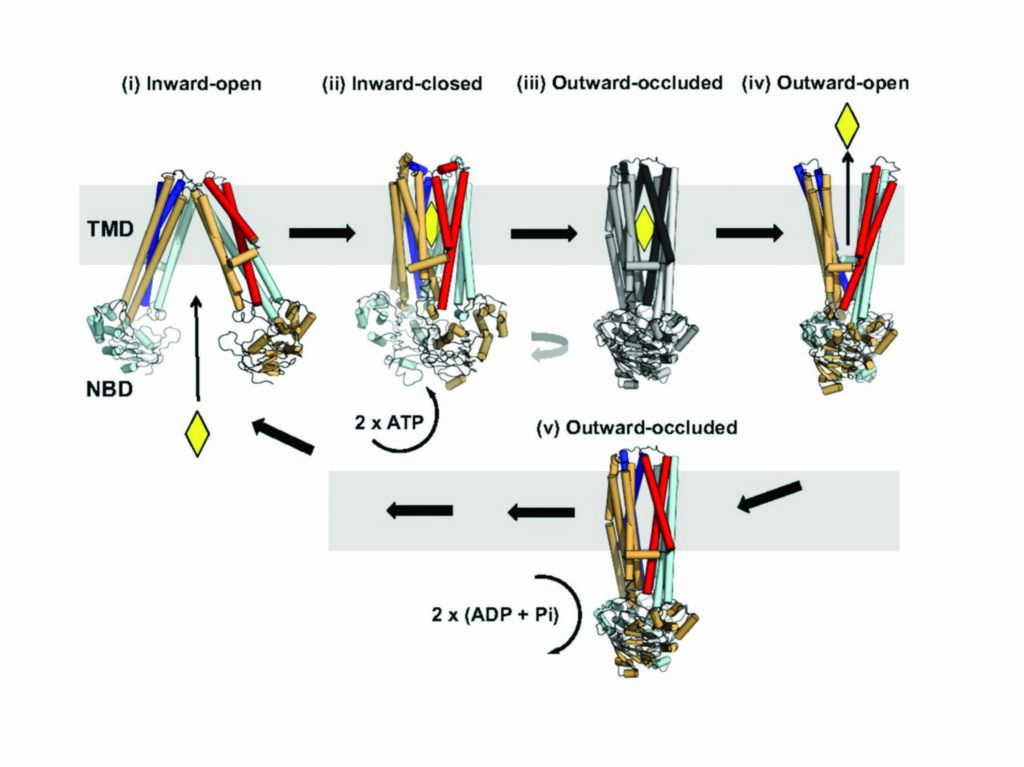
Figure 3: Mechanism of ABC exporters. Ligand binding (yellow diamond) to the inward-open conformation (state i) leads to the inward-closed conformation (state ii). Binding of ATP results in a transient outward occluded conformation (hypothetical state iii) and release of the substrate by the formation of an outward-open conformation (state iv). Once the substrate is released, the ABC exporter adopts an outward-occluded conformation (McjD structure, state v). The transporter resets back to an inward-facing conformation upon ATP hydrolysis.
Source publication:
Choudhury, H. G., Tong, Z., Mathavan, I., Li, Y., Iwata, S., Zirah, S., Rebuffat, S., van Veen, H. W. & Beis, K. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proceedings of the National Academy of Sciences of the United States of America 111, 9145-9150, doi:10.1073/pnas.1320506111 (2014).
References:
1. Duquesne, S., Destoumieux-Garzon, D., Peduzzi, J. & Rebuffat, S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Natural Product Reports 24, 708-734, doi:10.1039/b516237h (2007).
2. Choudhury, H. G. et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proceedings of the National Academy of Sciences of the United States of America 111, 9145- 9150, doi:10.1073/pnas.1320506111 (2014).
3. Dawson, R. J. P. & Locher, K. P. Structure of a bacterial multidrug ABC transporter. Nature 443, 180-185, doi:10.1038/nature05155 (2006).
4. Ward, A., Reyes, C. L., Yu, J., Roth, C. B. & Chang, G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proceedings of the National Academy of Sciences of the United States of America 104, 19005- 19010, doi:10.1073/pnas.0709388104 (2007).
Funding acknowledgements:
This research was funded by the Biotechnology and Biological Sciences Research Council (Grants BB/H01778X/1 to K.B., BB/G023425/1 to S.I., and BB/I002383/1 to H.W.v.V.) and Wellcome Trust (MPL: WT/099165/Z/12/Z to S.I.).
Corresponding author:
Dr Konstantinos Beis, Imperial College London, kbeis@imperial.ac.uk


 A brighter light for science
A brighter light for science
