 | ||
Structural and functional studies of lipopolysaccharide transport and insertion into the outer membrane by LptD/E complex |
Beamline I24 Scientific Highlight
In this research, a complex of two transport proteins, LptD and LptE, found in the outer membrane of a class of bacteria called Gram-negative bacteria, was studied. This class of bacteria have a unique outer membrane that is particularly difficult for antibiotics and other drugs to penetrate. To determine the structure of this complex and better understand the mechanism by which this protein transports LPS across the bacterial membrane, the crystal structure of the LptD/E complex was solved using Multi-wavelength Anomalous Dispersion (MAD) data on Macromolecular Crystallography (MX) beamline I24 with the aid of beamline staff. This approach relied on the small beam size and high flux density achievable by I24 to subdivide the crystal and collect 2.86 Å resolution data for each of the 4 wavelengths used. The Pilatus3 detector on I24 facilitated the collection of over 55,000 diffraction images across three crystals in a reasonable time frame that, when combined, allowed the generation of a high quality electron density map and subsequent protein model. The results have identified the path and gate that is used by the bacteria to transport LPS to the outer surface and more importantly that the bacteria would die if the gate was locked shut. These findings provide an exciting platform for future research to develop new compounds that target this specific system; potentially leading to the creation of drugs that bacteria will find it very difficult to become resistant to.
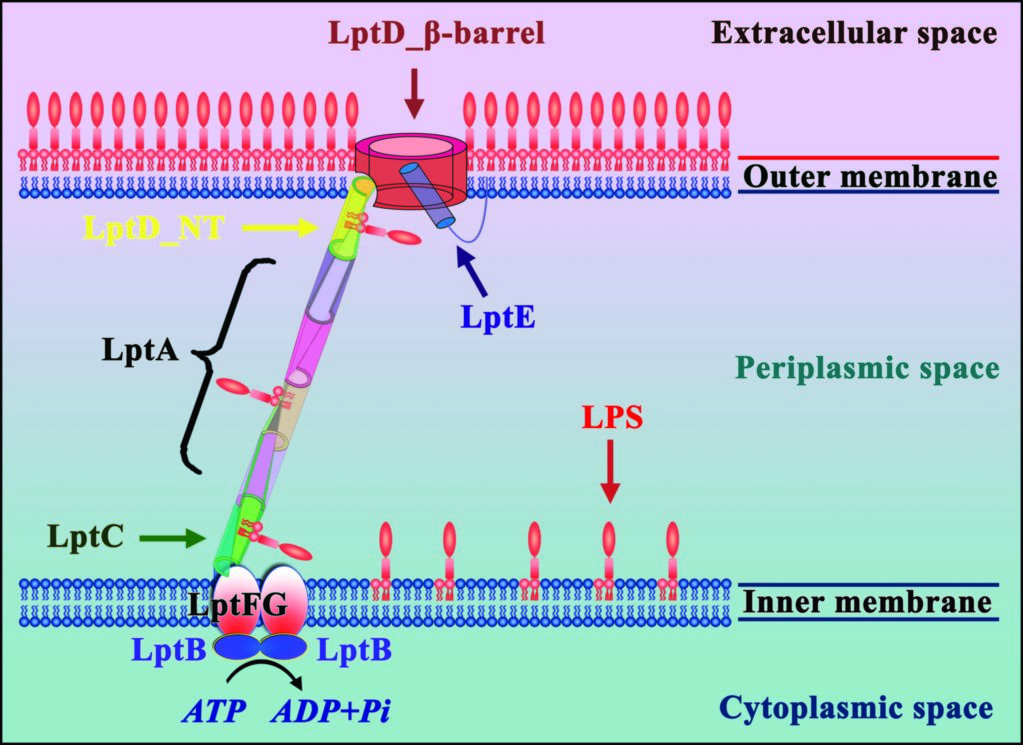
Figure 1: Seven LPS transport proteins form a trans-envelope complex to transport LPS from the inner membrane to the outer membrane.
All Gram-negative bacteria have an outer membrane, which comprises lipopolysaccharide (LPS) in the outer leaflet, and phospholipid in the inner leaflet. LPS is essential for most Gram-negative bacteria and plays a critical role in drug resistance. LPS contains three components: lipid A, core oligosaccharide, and O-antigen. The O-antigen is polymerised by WzY and ligated to the lipid A core oligosaccharide by WaaL1 to form the mature LPS on the exterior face of the inner membrane. Seven LPS transport proteins (LptA-LptG) have been identified as responsible for transport of the mature LPS from the inner membrane to the outer membrane (Fig. 1). Initially the inner membrane associated LptBFG complex uses the energy from ATP hydrolysis to transfer LPS from this membrane to LptC3. LPS is then guided across the bacterial periplasm on a ‘slide’ formed from LptC, LptA, and the N-terminal domain of LptD.
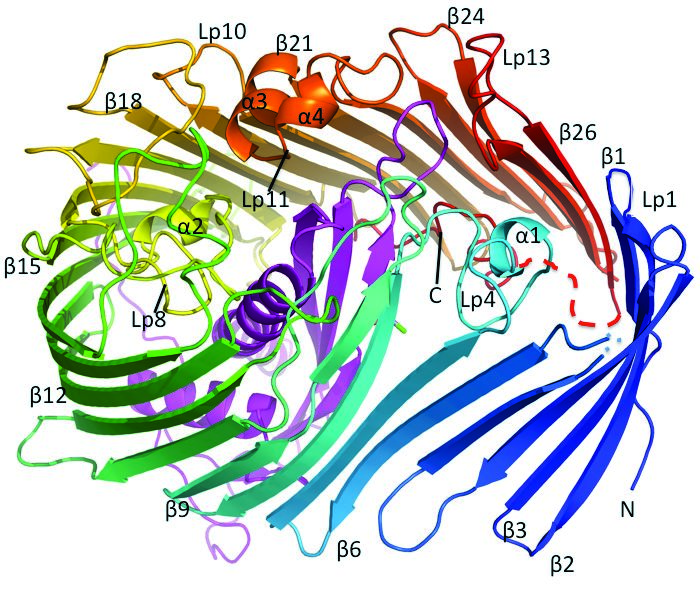
Figure 2: The S. typhimurium LT2 LptD/E complex structure. The LptD forms a 26 β-stranded barrel, while LptE is located inside of the barrel.
At the outset of this research, the mechanism by which the LptD/E complex translocates LPS across the outer membrane and inserts it into the outer leaflet was not understood. As the LptD/E complex is accessible from the exterior of the cell, it was conceivable that drugs targeting this complex may not need to enter the bacterial cell and thus sidestep both the LPS-rich outer membrane and also the efflux pumps that form key bacterial weapons in resisting antibiotics4. The research, aided by results gathered on I24, aimed to determine the structure of the S. typhimurium LT2 LptD/E complex structure and evaluate the novel LPS transport and insertion mechanism. Such structural information would also be invaluable for subsequent antibiotic development.
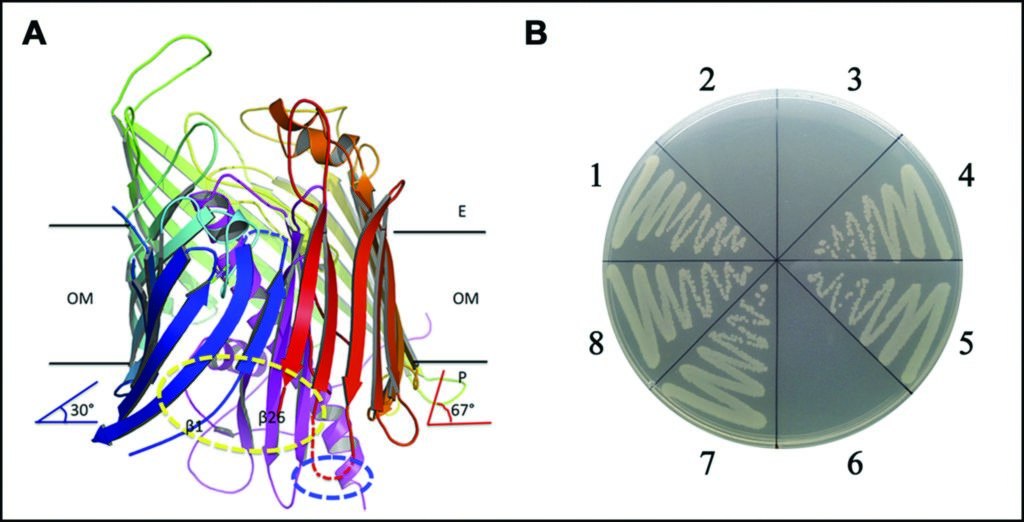
Figure 3: The separation between strands β1 and β26 at the periplasmic side, and possible lateral opening for LPS insertion. (a) the separation between strands β1 and β26 of LptD at the periplasmic side; (b) the functional assays for the double cysteine mutation.
Source publication:
Dong, H., Xiang, Q., Gu, Y., Wang, Z., Paterson, N. G., Stansfeld, P. J., He, C., Zhang, Y., Wang, W. & Dong, C. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52-56, doi:10.1038/nature13464 (2014).
References:
1. Raetz, C. R. H. & Whitfield, C. Lipopolysaccharide endotoxins. Annual Review of Biochemistry 71, 635-700, doi:10.1146/annurev. biochem.71.110601.135414 (2002).
2. Freinkman, E., Chng, S.-S. & Kahne, D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proceedings of the National Academy of Sciences of the United States of America 108, 2486-2491, doi:10.1073/pnas.1015617108 (2011).
3. Okuda, S., Freinkman, E. & Kahne, D. Cytoplasmic ATP Hydrolysis Powers Transport of Lipopolysaccharide Across the Periplasm in E-coli. Science 338, 1214-1217, doi:10.1126/science.1228984 (2012).
4. Srinivas, N. et al. Peptidomimetic Antibiotics Target Outer-Membrane Biogenesis in Pseudomonas aeruginosa. Science 327, 1010-1013, doi:10.1126/science.1182749 (2010)
Funding acknowledgement:
Wellcome Trust career development fellowship and Wellcome Trust New Investigator Award to CJD.
Corresponding author:
Professor Changjiang Dong, University of East Anglia, c.dong@uea.ac.uk


 A brighter light for science
A brighter light for science
