Summary
To contribute to the global effort to combat COVID-19, Diamond has been able to solve a new structure of the SARS-CoV-2 main protease (MPro) at high resolution (PDB ID: 6YB7), and complete a large XChem crystallographic fragment screen against it (detailed below). Data have been deposited with the PDB, but we are making the results available immediately to the world on this page; additional work is ongoing, and updates will be continually posted here in coming days and weeks.
This work builds on the sensationally fast crystal structure of MPro at 2.16 Å in complex with a covalent inhibitor, released in January this year by Prof Zihe Rao (6LU7, published here, described here). We thus ordered the synthetic gene and cloned the full length protein as previously described for the SARS main protease (Xue et al 2007). This yielded crystals of the unliganded enzyme that diffracted to high resolution (1.25 Å) on beamline I04-1, in a different space group to the inhibitor complex, and the structure was determined and refined rapidly. Critically, this showed it had the active site empty and solvent accessible - perfect for fragment screening.
So it proved: the first 600-crystal experiment could be completed in 72 hours, through growing large numbers of crystals, optimising the soaking conditions, soaking and harvesting all 600 crystals and completing the data collection run on beamline I04-1. The hits from this initial run and other details were pre-released on March 6th.
By the 24th of March, the initial 1500-crystal experiment was complete, and the results made publicly available. Screening additional libraries throughout April brought the total number of active site fragments to 71, with 48 fragments binding covalently (full timeline here and download page here). This was an exceptionally large screen which yielded a remarkably rich readout, with vast opportunities for fragment growing and merging.
We have already triggered computationally-driven follow-up work internally, and externally joined forces to launch a fully-open crowdsourcing and crowdfunding initiative – the COVID Moonshot - to establish urgently the shortest route possible to clinical impact by maximally exploiting the readout - you can help, read more here.
On the 11th of May, the first biochemical and structural data from Moonshot compounds was released and by the 12th of June over 500 compounds had been tested, demonstrating that the design-make-test process is fully in place.
XChem fragment screen
The initial screen encompassed multiple fragment libraries: the DSI-poised library, MiniFrags (Astex) FragLites & Peplites (CRUK Newcastle Drug Discovery Unit (Newcastle University)), York3D (University of York), SpotFinder and heterocyclic electrophilic fragment library (Hungarian Academy of Sciences) and an electrophilic fragment library designed and pre-screened by mass spec at the Weizmann Institute (see below).
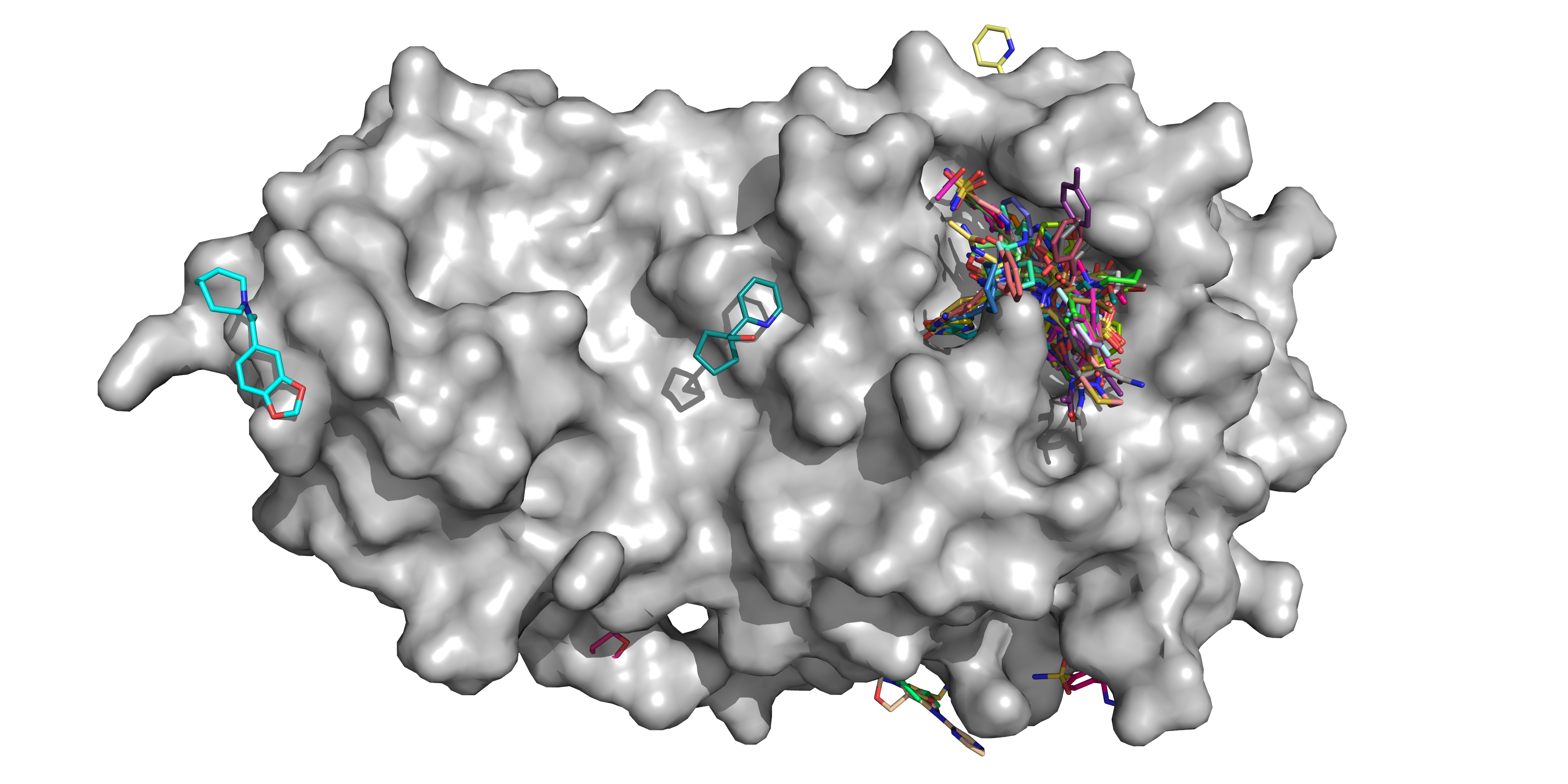
There were 74 hits of high interest - data and extensive details are here, and some interactive views here:
- 23 non-covalent hits in the active site
- 48 covalent hits in the active site
- 3 hits in the dimer interface, one in a calculated hotspot
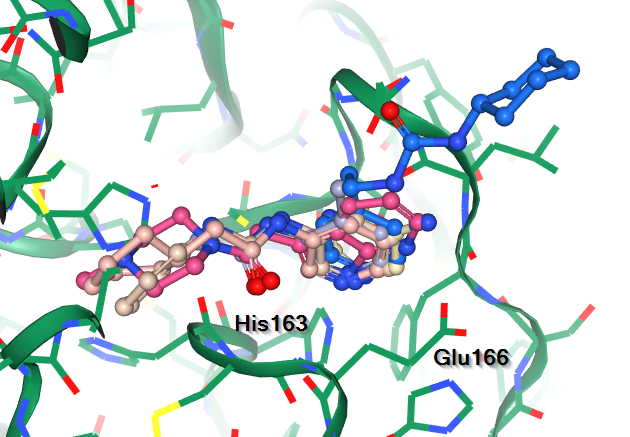
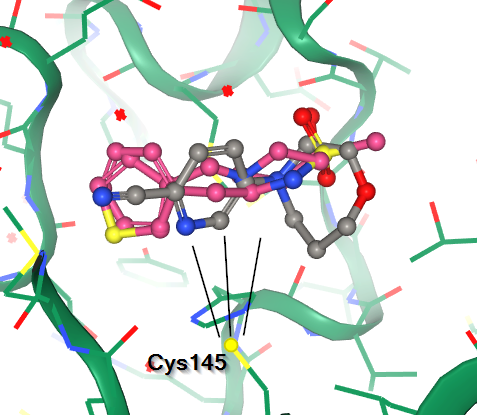
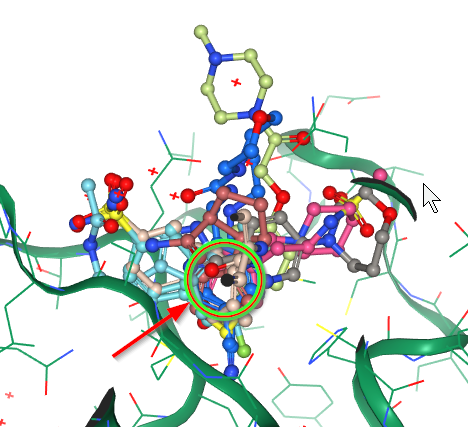
Features of note include:
| S1 - the His163 - Glu166 motif | S2 - hits over catalytic Cys145 | S3 - the aromatic wheel |
|
|
|
|
Mass spectrometry screening of electrophiles
The London lab screened Mpro supplied by Diamond with 993 fragment electrophiles by their intact-protein mass-spectrometry platform (Resnick et al. JACS 2019). A preliminary screen (200 µM, 24 h, 4°C) revealed 130 compounds that irreversibly label the protein (>50%); a more stringent screen (5 µM, 1.5 h, RT) discriminated several compound series that still label robustly:
The full experiment is summarised in this spreadsheet: 74 compounds were screened at XChem by crystal soaking and co-crystallisation following Resnick et al, working in triplicate: resulting in 42 complex crystal structures. (Of the rest, 29 yielded crystals without bound ligand, and 3 could not be measured for experimental reasons.)
A particularly promising series are the chloroacetamides based on piperidine-amides: as they were not hits in many previous screens, they should be very selective to Mpro. Additionally, they reveal SAR (Structure activity relationships) even amongst the nine representatives in the library, suggesting binding can be optimised. Based on the several crystal structures of these hits, we have commenced synthesis of analogs and testing of optimised derivatives.
Content last updated 5th of May, 2020
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.