Kamel El Omari
- Structural Biology
- Experimental/native phasing
- Viral proteins
Current Research Interests
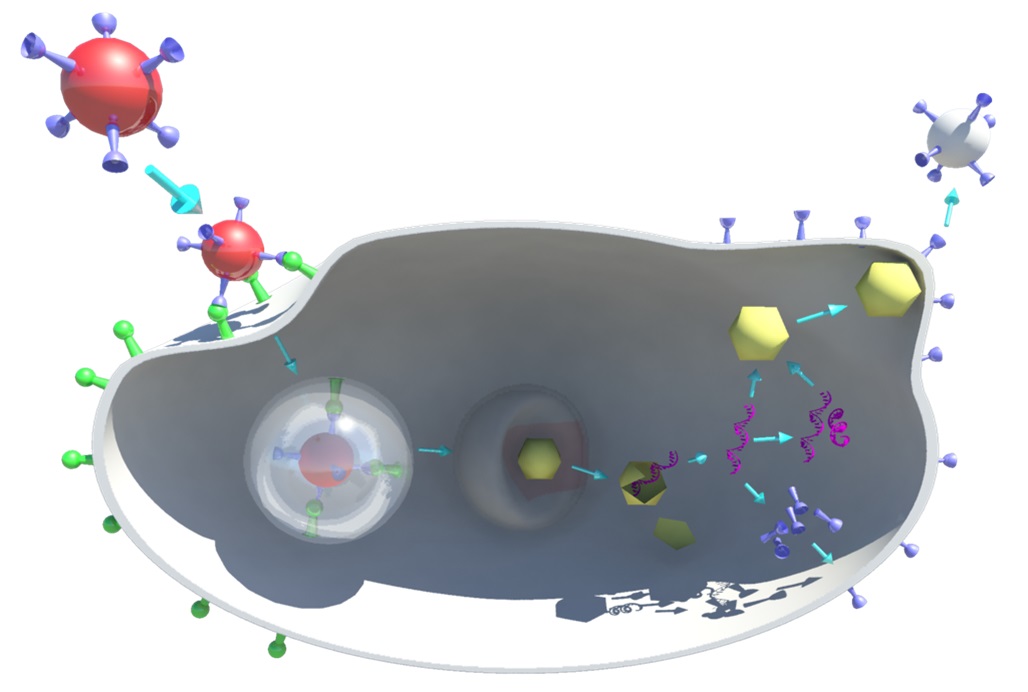
- Viral life cycle
Structural biology of viral membrane fusion
The aim of my project is to shed light on proteins interactions occurring between viruses and their host cells. Viruses use host cell resources to carry through their life cycle, but infection triggers host cell antiviral defense mechanisms that in turn drive viral adaptation. Thus it is crucial to understand not only the biology of the virus but also the biology of the host and the different cell pathways hijacked by the virus. This project will initially focus on unravelling the mechanism of viral membrane fusion.
HRPV6 virus
Native phasing
Single-wavelength anomalous dispersion of S atoms (S-SAD) is an elegant phasing method to determine crystal structures that does not require heavy-atom incorporation or selenomethionine derivatization. Nevertheless, this technique has been limited by the paucity of the signal at the usual X-ray wavelengths, requiring very accurate measurement of the anomalous differences. I23 has been designed to provide an optimized environment for difficult S-SAD experiments, by increasing the anomalous signal with minimal noise.
Publications
Duman R, Orr CM, Mykhaylyk V, El Omari K, Pocock R, Grama V, Wagner A. Sample Preparation and Transfer Protocol for In-Vacuum Long-Wavelength Crystallography on Beamline I23 at Diamond Light Source. J Vis Exp. (2021)
Riedel C, Aitkenhead H, El Omari K, Rümenapf T. Atypical Porcine Pestiviruses: Relationships and Conserved Structural Features. Viruses. (2021)
El Omari K, Mohamad N, Bountra K, Duman R, Romano M, Schlegel K, Kwong HS, Mykhaylyk V, Olesen C, Moller JV, Bublitz M, Beis K, Wagner A. Experimental phasing with vanadium and application to nucleotide-binding membrane proteins. IUCrJ. (2020)
Zhang Y, El Omari K, Duman R, Liu S, Haider S, Wagner A, Parkinson GN, Wei D. Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths. Nucleic Acids Res. (2020).
Mishra AK, Moyer CL, Abelson DM, Deer DJ, El Omari K, Duman R, Lobel L, Lutwama JJ, Dye JM, Wagner A, Chandran K, Cross RW, Geisbert TW, Zeitlin L, Bornholdt ZA, McLellan JS. Structure and Characterization of Crimean-Congo Hemorrhagic Fever Virus GP38. J Virol. (2020)
Qu F, ElOmari K, Wagner A, De Simone A, Beis K. Desolvation of the substrate-binding protein TauA dictates ligand specificity for the alkanesulfonate ABC importer TauABC. Biochem J. (2019)
Rocchio S, Duman R, El Omari K, Mykhaylyk V, Orr C, Yan Z, Salmon L, Wagner A, Bardwell JCA, Horowitz S. Identifying dynamic, partially occupied residues using anomalous scattering. Acta Crystallogr D Struct Biol. (2019)
Rudolf AF, Kinnebrew M, Kowatsch C, Ansell TB, El Omari K, Bishop B, Pardon E, Schwab RA, Malinauskas T, Qian M, Duman R, Covey DF, Steyaert J, Wagner A, Sansom MSP, Rohatgi R, Siebold C. The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism. Nat Chem Biol. (2019)
Rozov A, Khusainov I, El Omari K, Duman R, Mykhaylyk V, Yusupov M, Westhof E, Wagner A, Yusupova G. Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction. Nat Commun. (2019)
Ilca SL, Sun X, El Omari K, Kotecha A, de Haas F, DiMaio F, Grimes JM, Stuart DI, Poranen MM, Huiskonen JT. Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus. Nature (2019)
El Omari K, Li S, Kotecha A, Walter TS, Bignon EA, Harlos K, Somerharju P, De Haas F, Clare DK, Molin M, Hurtado F, Li M, Grimes JM, Bamford DH, Tischler ND, Huiskonen JT, Stuart DI, Roine E. The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins. Nat Commun. (2019)
Langan PS, Vandavasi VG, Weiss KL, Afonine PV, El Omari K, Duman R, Wagner A, Coates L. Anomalous X-ray diffraction studies of ion transport in K+ channels. Nat Commun. (2018)
Pryce R, Ng WM, Zeltina A, Watanabe Y, El Omari K, Wagner A, Bowden TA. Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1. J Virol. (2018)
Austin HP, Allen MD, Donohoe BS, Rorrer NA, Kearns FL, Silveira RL, Pollard BC, Dominick G, Duman R, El Omari K, Mykhaylyk V, Wagner A, Michener WE, Amore A, Skaf MS, Crowley MF, Thorne AW, Johnson CW, Woodcock HL, McGeehan JE, Beckham GT. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc Natl Acad Sci (2018)
Esposito D, Günster RA, Martino L, El Omari K, Wagner A, Thurston TLM, Rittinger K. Structural basis for the glycosyltransferase activity of the Salmonella effector SseK3. J Biol Chem. (2018)
Bountra K, Hagelueken G, Choudhury HG, Corradi V, El Omari K, Wagner A, Mathavan I, Zirah S, Yuan Wahlgren W, Tieleman DP, Schiemann O, Rebuffat S, Beis K. Structural basis for antibacterial peptide self-immunity by the bacterial ABC transporter McjD. EMBO J (2017)
Zhaoyang Sun, Kamel El Omari, Xiaoyu Sun, Serban L Ilca, Abhay Kotecha, David I Stuart, Minna M Poranen, Juha T Huiskonen. Double-stranded RNA virus outer shell assembly by bona fide domain-swapping. Nature Communications (2017).
Sina Wittmann, Max Renner, Beth R. Watts, Oliver Adams, Miles Huseyin, Carlo Baejen, Kamel El Omari, Cornelia Kilchert, Dong-Hyuk Heo, Tea Kecman, Patrick Cramer, Jonathan M. Grimes & Lidia Vasiljeva.The conserved protein Seb1 drives transcription termination by binding RNA polymerase II and nascent RNA. Nature Communications (2017).
Oskar Aurelius, Ramona Duman, Kamel El Omari, Vitaliy Mykhaylyk, Armin Wagner. Long-wavelength macromolecular crystallography–First successful native SAD experiment close to the sulfur edge. NIMB (2016).
Thierry, Eric; Guilligay, Delphine; Kosinski, Jan; Bock, Thomas; Gaudon, Stephanie; Round, Adam; Pflug, Alexander; Hengrung, Narin; El Omari, Kamel; Baudin, Florence. Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of PB2 Domains. Molecular cell 61 (1), 125-137, 2016.
Vendelboe, Trine V; Harris, Pernille; Zhao, Yuguang; Walter, Thomas S; Harlos, Karl; El Omari, Kamel; Christensen, Hans EM. The crystal structure of human dopamine β-hydroxylase at 2.9 Å resolution. Science Advances 2 (4), e1500980, 2016.
Lye, Ming F; Sharma, Mayuri; El Omari, Kamel; Filman, David J; Schuermann, Jonathan P; Hogle, James M; Coen, Donald M. Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex. The EMBO journal 34 (23), 2937-2952, 2015.
Hengrung, Narin; El Omari, Kamel; Martin, Itziar Serna; Vreede, Frank T; Cusack, Stephen; Rambo, Robert P; Vonrhein, Clemens; Bricogne, Gérard; Stuart, David I; Grimes, Jonathan M. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus, Nature 527 (7576), 114-117, 2015.
Zeev-Ben-Mordehai, Tzviya; Weberruß, Marion; Lorenz, Michael; Cheleski, Juliana; Hellberg, Teresa; Whittle, Cathy; El Omari, Kamel; Vasishtan, Daven; Dent, Kyle C; Harlos, Karl. Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling. Cell reports 13 (12), 2645-2652, 2015.
Sewell, H; Tanaka, T; El Omari, Kamel; Mancini, EJ; Cruz, A; Fernandez-Fuentes, N; Chambers, J; Rabbitts, TH. Conformational flexibility of the oncogenic protein LMO2 primes the formation of the multi-protein transcription complex. Scientific reports 4, 2014.
El Omari, Kamel; Iourin, Oleg; Kadlec, Jan; Fearn, Richard; Hall, David R; Harlos, Karl; Grimes, Jonathan M; Stuart, David I. Pushing the limits of sulfur SAD phasing: de novo structure solution of the N-terminal domain of the ectodomain of HCV E1. Acta D 70 (8) 2197-2203, 2014.
El Omari, Kamel; Iourin, Oleg; Kadlec, Jan; Sutton, Geoff; Harlos, Karl; Grimes, Jonathan M; Stuart, David I. Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nature communications 5, 2014.
Iourin, Oleg; Harlos, Karl; El Omari, Kamel; Lu, Weixian; Kadlec, Jan; Iqbal, Munir; Meier, Christoph; Palmer, Andrew; Jones, Ian; Thomas, Carole. Expression, purification and crystallization of the ectodomain of the envelope glycoprotein E2 from Bovine viral diarrhoea virus. Acta F 69 (1) 35-38, 2013.
El Omari, Kamel; Iourin, Oleg; Harlos, Karl; Grimes, Jonathan M; Stuart, David I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell reports 3 (1) 30-35, 2013.
Nichols, CE; Lamb, HK; Thompson, P; El Omari, Kamel; Lockyer, M; Charles, I; Hawkins, AR; Stammers, DK. Crystal structure of the dimer of two essential Salmonella typhimurium proteins, YgjD & YeaZ and calorimetric evidence for the formation of a ternary YgjD–YeaZ–YjeE complex. Protein Science 22 (5) 628-640, 2013.
El Omari, Kamel; Hoosdally, Sarah J; Tuladhar, Kapil; Karia, Dimple; Hall-Ponselé, Elisa; Platonova, Olga; Vyas, Paresh; Patient, Roger; Porcher, Catherine; Mancini, Erika J. Structural Basis for LMO2-Driven Recruitment of the SCL: E47 bHLH Heterodimer to Hematopoietic-Specific Transcriptional Targets. Cell reports 4 (1) 135-147, 2013.
El Omari, Kamel; Sutton, Geoff; Ravantti, Janne J; Zhang, Hanwen; Walter, Thomas S; Grimes, Jonathan M; Bamford, Dennis H; Stuart, David I; Mancini, Erika J. Plate tectonics of virus shell assembly and reorganization in phage φ8, a distant relative of mammalian reoviruses. Structure 21 (8) 1384-1395, 2013.
El Omari, Kamel; Meier, Christoph; Kainov, Denis; Sutton, Geoff; Grimes, Jonathan M; Poranen, Minna M; Bamford, Dennis H; Tuma, Roman; Stuart, David I; Mancini, Erika J. Tracking in atomic detail the functional specializations in viral RecA helicases that occur during evolution. Nucleic acids research 41 (20) 9396-9410, 2013.
Bourhis, J-M; Mariano, N; Zhao, Y; Walter, TS; El Omari, Kamel; Delolme, F; Moali, C; Hulmes, DJS; Aghajari, N. Production and crystallization of the C-propeptide trimer from human procollagen III. Acta F 68 (10) 1209-1213, 2012.
El Omari, Kamel; De Mesmaeker, Julie; Karia, Dimple; Ginn, Helen; Bhattacharya, Shoumo; Mancini, Erika J. Structure of the DNA‐bound T‐box domain of human TBX1, a transcription factor associated with the DiGeorge syndrome. Proteins: Structure, Function, and Bioinformatics 80 (2) 655-660, 2012.
El Omari, Kamel; Hoosdally, Sarah J; Tuladhar, Kapil; Karia, Dimple; Vyas, Paresh; Patient, Roger; Porcher, Catherine; Mancini, Erika J. Structure of the leukemia oncogene LMO2: implications for the assembly of a hematopoietic transcription factor complex. Blood 117 (7) 2146-2156, 2011.
Stamp, Anna L; Owen, Paul; El Omari, Kamel; Lockyer, Michael; Lamb, Heather K; Charles, Ian G; Hawkins, Alastair R; Stammers, David K. Crystallographic and microcalorimetric analyses reveal the structural basis for high arginine specificity in the Salmonella enterica serovar Typhimurium periplasmic binding protein STM4351. Proteins: Structure, Function, and Bioinformatics 79 (7) 2352-2357, 2011.
El Omari, Kamel; Dhaliwal, B; Ren, J; Abrescia, NGA; Lockyer, M; Powell, KL; Hawkins, AR; Stammers, DK. Structures of respiratory syncytial virus nucleocapsid protein from two crystal forms: details of potential packing interactions in the native helical form. Acta F 67 (10) 1179-1183, 2011.
Stamp, Anna L; Owen, Paul; El Omari, Kamel; Nichols, Charles E; Lockyer, Michael; Lamb, Heather K; Charles, Ian G; Hawkins, Alastair R; Stammers, David K. Structural and functional characterization of Salmonella enterica serovar Typhimurium YcbL: an unusual Type II glyoxalase. Protein Science 19 (10) 1897-1905, 2010.
El Omari, Kamel; Porcher, Catherine; Mancini, Erika J. Purification, crystallization and preliminary X-ray analysis of a fusion of the LIM domains of LMO2 and the LID domain of Ldb1. Acta F 66 (11) 1466-1469, 2010.
El Omari, Kamel; Scott, K; Dhaliwal, B; Ren, J; Abrescia, NGA; Budworth, J; Lockyer, M; Powell, KL; Hawkins, AR; Stammers, DK. Crystallization and preliminary X-ray analysis of the human respiratory syncytial virus nucleocapsid protein. Acta F 64 (11) 1019-1023, 2008.
Szumska, Dorota; Pieles, Guido; Essalmani, Rachid; Bilski, Michal; Mesnard, Daniel; Kaur, Kulvinder; Franklyn, Angela; El Omari, Kamel; Jefferis, Joanna; Bentham, Jamie. VACTERL/caudal regression/Currarino syndrome-like malformations in mice with mutation in the proprotein convertase Pcsk5. Genes & development 22 (11) 1465-1477, 2008.
El Omari, Kamel; Stammers, David K. The design and development of drugs acting against the smallpox virus. Expert opinion on drug discovery 2 (9) 1263-1272, 2007.
El Omari, Kamel; Ren, Jingshan; Bird, Louise E; Bona, Marion K; Klarmann, George; LeGrice, Stuart FJ; Stammers, David K. Molecular architecture and ligand recognition determinants for T4 RNA ligase. JBC 281 (3) 1573-1579, 2006.
El Omari, Kamel; Solaroli, Nicola; Karlsson, Anna; Balzarini, Jan; Stammers, David K. Structure of vaccinia virus thymidine kinase in complex with dTTP: insights for drug design. BMC structural biology 6 (1) 1, 2006.
Balzarini, Jan; Liekens, Sandra; Solaroli, Nicola; El Omari Kamel; Stammers, David K; Karlsson, Anna. Engineering of a single conserved amino acid residue of herpes simplex virus type 1 thymidine kinase allows a predominant shift from pyrimidine to purine nucleoside phosphorylation. JBC 281 (28) 19273-19279, 2006.
El Omari, Kamel; Liekens, Sandra; Bird, Louise E; Balzarini, Jan; Stammers, David K. Mutations distal to the substrate site can affect varicella zoster virus thymidine kinase activity: implications for drug design. Molecular pharmacology 69 (6) 1891-1896, 2006.
El Omari, Kamel; Dhaliwal, Balvinder; Lockyer, Michael; Charles, Ian; Hawkins, Alastair R; Stammers, David K; Structure of Staphylococcus aureus guanylate monophosphate kinase. Acta F 62 (10) 949-953, 2006.
El Omari, Kamel; Bronckaers, Annelies; Liekens, Sandra; Pérez-Pérez, Maris-Jésus; Balzarini, Jan; Stammers, David K. Structural basis for non-competitive product inhibition in human thymidine phosphorylase: implications for drug design. Biochemical Journal 399 (2) 199-204, 2006.
El Omari, Kamel; Bird, Louise E; Nichols, Charles E; Ren, Jingshan; Stammers, David K. Crystal structure of CC3 (TIP30) implications for its role as a tumor suppressor. JBC 280 (18) 18229-18236, 2005.


 Macromolecular Crystallography
Macromolecular Crystallography

