A pivotal player in antibiotic resistance, known as MCR-1, has recently been characterised at Diamond Light Source. The crystal structure and mechanistic insights from this study have been published in Scientific Reports and they will likely contribute to developments to stave off the threat of antibiotic resistance.
Antibiotic resistance is a global concern and a top health priority in many countries. Resistance in Gram-negative bacteria, such as Escherichia coli and Klebsiella pneumoniae, is a particular worry as they are responsible for a slew of common infections that could potentially lead to death if they are not controlled effectively. Recently, a plasmid encoding a resistance factor known as MCR-1 was discovered in these two infectious bacteria in China.
MCR-1 is an enzyme that allows bacteria to become resistant to a key antibiotic known as colistin. MCR-1 modifies the binding site for colistin to render it ineffective, but the molecular mechanisms underpinning this interaction are as yet unknown. Using the Macromolecular Crystallography (MX) beamlines I02, I03 and I04-1 at Diamond, an international collaboration of scientists demonstrated that the catalytic domain of the enzyme was a zinc metalloprotein with an alkaline phosphatase/sulphatase fold containing three disulphide bonds. The team also predicted the mechanism of action of the enzyme, concluding that a single zinc ion was sufficient for the catalytic reaction.
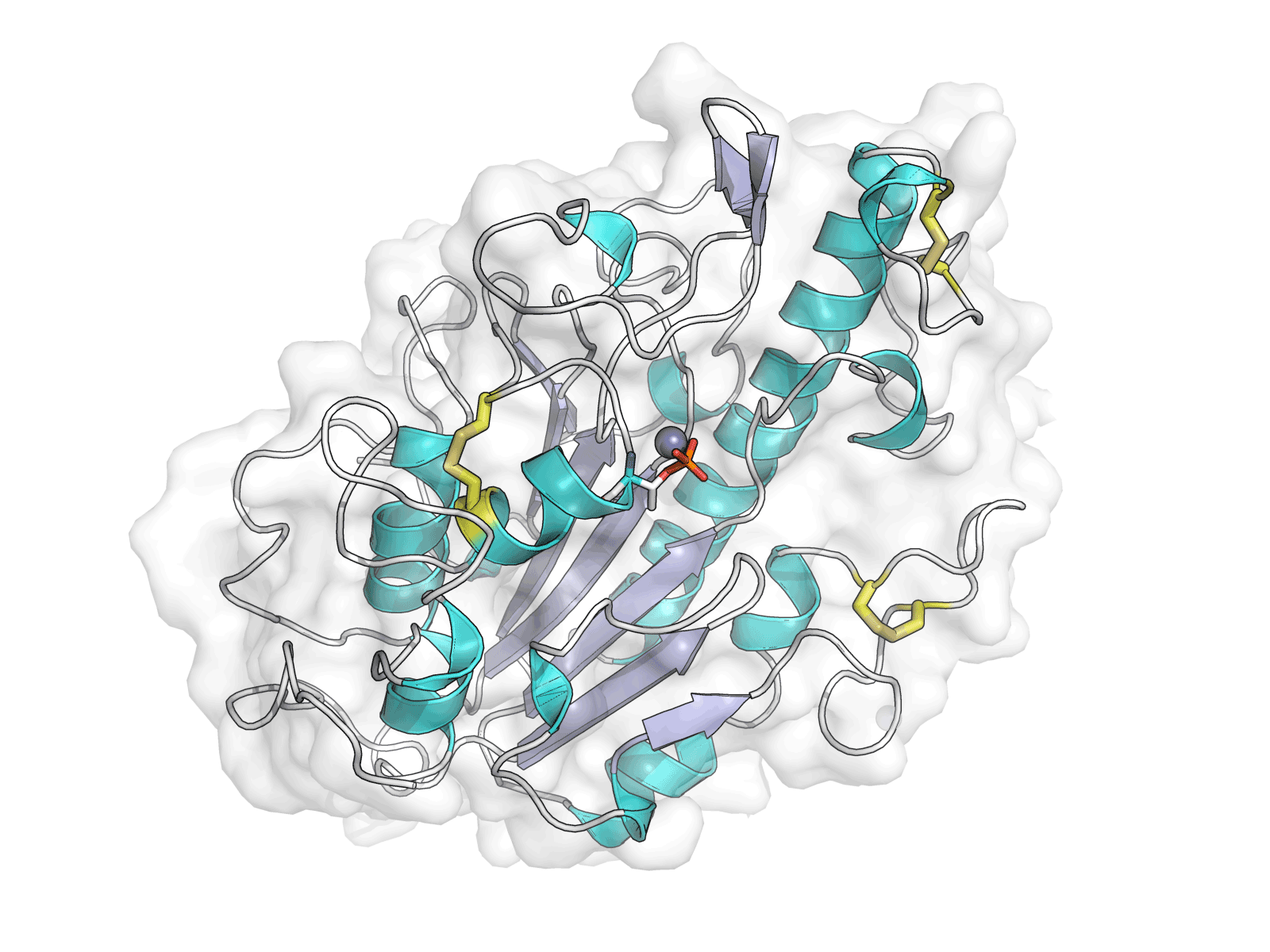
Hinchliffe P et al. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci Rep. 7, 39392 (2017). DOI: 10.1038/srep39392
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.