___________________________________
Industrial Liaison Group:
Tel: +44 (0) 1235 778797
E-mail: industry@diamond.ac.uk
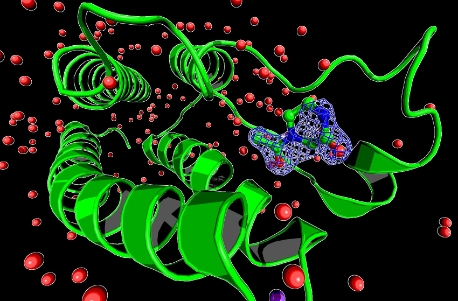
Scientists have utilised Diamond Light Source to develop a new method to extract previously hidden information from the X-ray diffraction data that are measured when resolving the three-dimensional (3D) atomic structures of proteins and other biological molecules.
When trying to evolve chemical compounds towards potent drug candidates, scientists attempt to study the atomic detail of how compounds bind to their target proteins. To do so, they compare X ray data measured in both presence and absence of the compound. However, with existing analysis algorithms, this difference signal can often be swamped by noise from experiment artefacts, making it very unreliable to interpret the observed signal.
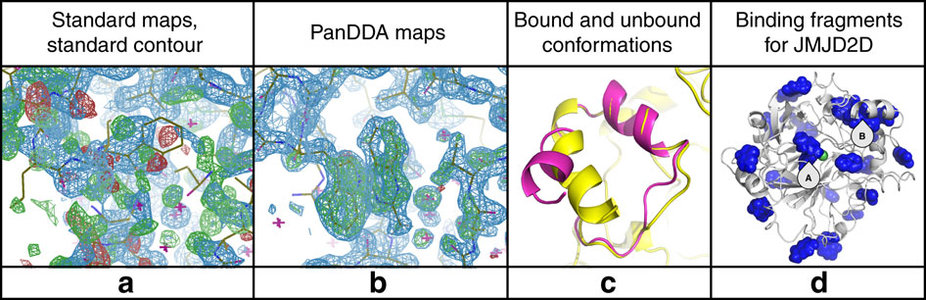
The new Pan-Dataset Density Analysis (PanDDA) method extracts the picture of the bound compound in exceptionally clear and unambiguous detail. PanDDA first identifies the source of the noise, and then removes it from the data. It exploits Diamond’s ability to repeat dozens to hundreds of measurements quickly, which are then characterised for differences between them, indicating the presence of bound compound, after which a noise correction is applied in 3D. This powerful algorithm is now an integral part of Diamond’s XChem platform and is routinely used on fragment screening campaigns to rapidly extract fragment hits.
The results have been recently published in Nature Communications. Macromolecular crystallography (MX), the technique that PanDDA applies to, is one of the most powerful tools used by researchers interested in determining the 3D structures of large biological molecules, including proteins, and is the work-horse experiment for rational drug design.
 “The problem of identifying binding events in crystallographic datasets can feel like looking for a needle in a haystack,” explains Dr Nicholas Pearce, lead author on the paper which comes from his PhD project at the University of Oxford in the Systems Approaches for Biomedical Science (SABS) Centre for Doctoral Training, where he was jointly funded by UCB Pharma and Diamond. “In the case of the data we were analysing, it was even worse, because we had hundreds of haystacks, and didn’t know which of them contained needles.” Dr Pearce is now based in the Crystal & Structural Chemistry Group at Universiteit Utrecht.
“The problem of identifying binding events in crystallographic datasets can feel like looking for a needle in a haystack,” explains Dr Nicholas Pearce, lead author on the paper which comes from his PhD project at the University of Oxford in the Systems Approaches for Biomedical Science (SABS) Centre for Doctoral Training, where he was jointly funded by UCB Pharma and Diamond. “In the case of the data we were analysing, it was even worse, because we had hundreds of haystacks, and didn’t know which of them contained needles.” Dr Pearce is now based in the Crystal & Structural Chemistry Group at Universiteit Utrecht.“UCB is delighted to have been working closely with Diamond on the development of PanDDA and its application to crystallographic fragment screening,” comments Dr Neil Weir, Senior Vice President of Discovery at UCB Pharma. “As a direct result, we have been able to identify fragments, which were otherwise not distinguishable from background, bound to a key protein-protein interaction drug target.”
Would you like to know more about PanDDA and how you can apply it to your research? Then please get in touch with the Industrial Liaison Team at Diamond.
The Industrial Liaison team at Diamond is a group of professional, experienced scientists with a diverse range of expertise, dedicated to helping scientists and researchers from industry access the facilities at Diamond. We’re all specialists in different techniques and have a diverse range of backgrounds so we’re able to provide a multi-disciplinary approach to solving your research problems.
Pearce, N. M. et al. A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat. Commun. 8, 15123 doi: 10.1038/ncomms15123 (2017), is available at: www.nature.com/articles/ncomms15123.
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.