The Hsf protein found in virulent strains of the bacteria Haemophilus influenzae is thought to aid the pathogen avoiding our immune response, leading to diseases such as pneumonia and meningitis. In a paper published in Acta Cryst. F, Professor Adrian Goldman’s team at the University of Leeds used the Macromolecular Crystallography beamline (I03) at Diamond Light Source to determine the structure of a subdomain of Hsf known as PD1, revealing a structural component missed by computational predictions. Understanding the PD1 structure brings the team one step closer to uncovering the structure of the entire protein, which could then be used to design molecular antagonists that would block Haemophilus influenzae’s evasion of the immune system.
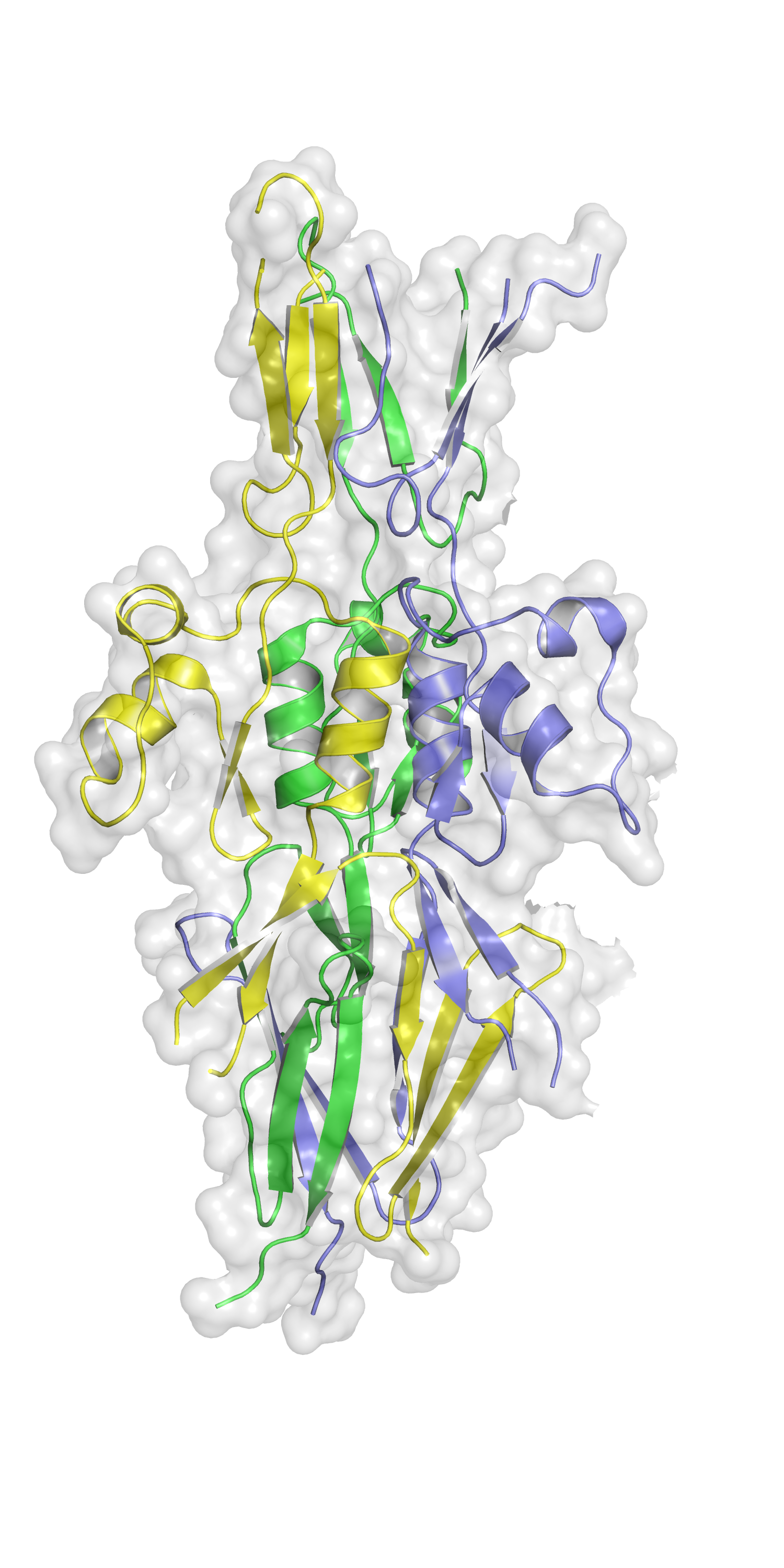
The PD1 structure is just one step along the road. The team are planning further experiments at Diamond and using cryo-electron microscopy to unveil the structure of the entire Hsf protein. This will enable them to determine how the protein presents on Haemophilus influenzae and which portion contains the binding site. Prof Goldman hopes to discover what immune molecules Hsf binds and find a way to interfere with this process. “Instead of trying to kill the bacteria, if we just prevent them shielding themselves from attack by the immune system, then the immune system will clean them up. That’s what I think the future of antibiotics looks like.”
To find out more about the I03 beamline, or to discuss potential applications, please contact Principal Beamline Scientist Dr Katherine McAuley: [email protected]
Wright J et al. The crystal structure of PD1, a Haemophilus surface fibril domain. Acta Crystallographica Section F 73, 101-108 (2017). DOI: 10.1107/S2053230X17001406.
Diamond Light Source is the UK's national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire.
Copyright © 2022 Diamond Light Source
Diamond Light Source Ltd
Diamond House
Harwell Science & Innovation Campus
Didcot
Oxfordshire
OX11 0DE
Diamond Light Source® and the Diamond logo are registered trademarks of Diamond Light Source Ltd
Registered in England and Wales at Diamond House, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0DE, United Kingdom. Company number: 4375679. VAT number: 287 461 957. Economic Operators Registration and Identification (EORI) number: GB287461957003.