Integrated Facilities
Membrane Protein Laboratory
Since completing its set up phase in 2010, the Membrane Protein Laboratory (MPL) has provided a research and training facility for scientists the world over interested in solving the 3D structures of membrane proteins by X-ray crystallography. Funded by the Wellcome Trust, it is the only laboratory of its kind to be situated just metres from a synchrotron radiation experimental station, Diamond’s Microfocus Macromolecular Crystallography (MX) beamline (I24).
The unique setup provided by the combination of the two facilities has led to a decrease in the time taken to solve the structure of a membrane protein, which is typically at a rate of one per every three years. Since 2012 the MPL has helped to solve one new membrane protein structure per year. One of the most recent structures was reported in Nature Structural & Molecular Biology, revealing how sodium is transported into and out of our cells (Fig. 1)1. This detailed knowledge could have important implications for health problems related to cancer and hypertension.
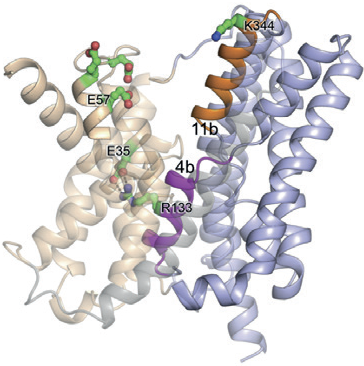 Figure 1: Ribbon representation of NapA (blue, core; wheat, dimer) in inward-facing1.
Figure 1: Ribbon representation of NapA (blue, core; wheat, dimer) in inward-facing1.
In line with the new facilities available at Diamond, the MPL is developing expertise within electron microscopy (EM) and X-ray free electron laser (XFEL) experiments. Following equipment upgrades, the MPL will offer training in these areas to help users prepare their samples for EM and XFEL experiments, as well as continuing its successful hands-on training programme in preparing for X-ray crystallography experiments. In 2015 Head of the MPL, Isabel de Moraes, took part in membrane protein experiments, along with the Structural Genomics Consortium (SGC) Oxford, at the Linac Coherent Light Source (LCLS), at Stanford University’s SLAC National Accelerator Laboratory, California, and SPring-8 Angstrom Compact Free Electron Laser (SACLA), Japan. SGC Oxford and the MPL are currently the only groups taking membrane proteins grown in the UK to XFEL beamlines. With the resources and expertise provided by the MPL, we expect to see a rapid growth in this area in years to come.
Overall the MPL provides support to a significant, and still growing, number of users from the UK and overseas. In the last five years more than 120 applications from 21 different institutions have been submitted to the MPL. This number is set to increase as the lab continues to perform beyond its remit; delivering double the number of projects set out for the year.
References:
- M. Coincon, P. Uzdavinys, E. Nji, D. L. Dotson, I. Winkelmann, S. Abdul-Hussein, A. D. Cameron, O. Beckstein and D. Drew. Crystal structures reveal the molecular basis of ion translocation in sodium/ proton antiporters. Nature Structural & Molecular Biology 23, 248-255
doi: 10.1038/nsmb.3164 (2016).

Figure 2: Head of MPL, Isabel de Moraes, at the Linac Coherent Light Source (LCLS), California.
eBIC: electron Bio-Imaging Centre
Funded by a £15.6 million grant from the Wellcome Trust, the Medical Research Council (MRC), and the Biotechnology and Biological Sciences Research Council (BBSRC), eBIC is a collaboration between Diamond, Birkbeck College and the University of Oxford. With advanced microscopes, detectors and software, eBIC allows scientists to investigate and visualise the structure of individual cells and bio-molecules, complementing Diamond’s current capabilities.
eBIC’s first cryo-EM to come online is the Titan Krios I, which is currently housed within the Diamond synchrotron building. The microscope went into operation in June 2015, when Venki Ramakrishnan’s group from the MRC Laboratory of Molecular Biology used it to study the structure of the yeast mitochondrial ribosome. Since then the microscope has seen 66 user visits by 44 unique groups, including almost all of the established cryo-EM groups in the UK, with a large amount of interest coming from overseas groups, as well as less established groups. The second microscope will have the same specification as the first and is due to come online summer 2016.
Designed for rapid, stable, high-resolution data collection on frozenhydrated samples, the first microscope has already yielded ground breaking results. The findings published in journals Cell and Nature Communications ultimately have the potential to support new drugs and therapies.
Battling urinary tract infections
The Cell paper describes how a team from University College London (UCL) and Birkbeck, University of London, the University of Washington, and the University of Virginia used the advanced microscope to study the bacterium responsible for causing urinary tract infections (UTIs). They focused on the bacterium’s tentacle-like appendages, known as pili, which play an essential role in infection as they are responsible for seeking out and latching onto specific host tissues.

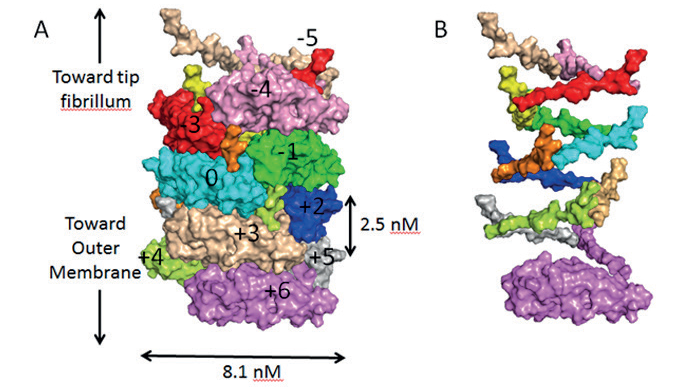
Figure 1: A) Surface diagram of the pilus in its coiled state, showing each subunit in a different colour. The subunit coloured cyan is the reference subunit and is numbered ‘0’. Subunits above or below this reference are numbered accordingly. B) Surface diagram of the pilus in its uncoiled state, showing only the N-terminal extension sections to clearly illustrate the helical organisation of the pilus.
The pili were made in vitro and applied to EM grids, which were flash frozen prior to analysis at eBIC where data was collected in two days, and the atomic structure was finalised two weeks later. The team were able to elucidate the structure of the pili found on E.coli and produce an atomic model generated from a 3.8 Å resolution cryo-EM reconstruction (Fig. 1).
The structural information revealed that the pili are coiled tightly, like a spring. When enduring the fierce currents of urine flow, the coiled pili flex and bend like an old-fashioned telephone cord, letting the bacteria withstand forces that otherwise would be strong enough to dislodge them and wash them away.
Never before has the structure of the P pilus been resolved, so this study has given scientists an important insight into mechanisms employed by bacteria to promote infection. By fully understanding the structure and function of these bacterial appendages, it is thought that new medicines could be developed to prevent infection.
The group next plan to explore how the pili are formed and intend to disrupt this process by introducing a series of mutations into the pili to potentially weaken their spring-like property and see how their growth is affected.
Related publication:
- Hospenthal MK et al. Structure of a chaperone-usher pilus reveals the molecular basis of rod uncoiling. Cell (2016)
doi:10.1016/j.cell.2015.11.049.
Punching holes in bacteria
The Nature Communications paper reports the findings of a team of scientists from Imperial College London and Cardiff University. They used the cryo-EM facilities at Diamond to reveal the structure of a fundamental part of our immune system, the membrane attack complex (MAC), a critical player in our response to infection. When pathogens are detected, a collection of signalling proteins are activated, triggering the assembly of the MAC, a huge pore that literally punches holes in the wall of bacteria to kill them. Although the lesions made by this protein complex were first noted in 1964, the actual structure of MAC was unknown until now. Thanks to the facilities and resources provided by eBIC, we now have an insight into how this unusual complex breaks through microbial membranes.
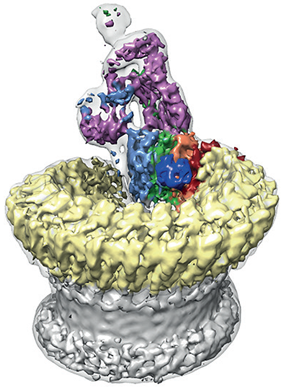 Figure 2: Sharpened map segmented and coloured according to complement proteins: C5b (magenta), C6 (light blue), C7 (green), C8a (red), C8b (orange), C8g (dark blue) and C9 (light yellow). CDC/MACPF β-barrel and detergent belt are in grey colour. Density is overlaid on the unsharpened map (transparent surface).
Figure 2: Sharpened map segmented and coloured according to complement proteins: C5b (magenta), C6 (light blue), C7 (green), C8a (red), C8b (orange), C8g (dark blue) and C9 (light yellow). CDC/MACPF β-barrel and detergent belt are in grey colour. Density is overlaid on the unsharpened map (transparent surface).
The team found that the pore has a split washer shape (Fig. 2). Unlike closely related pore-forming proteins that form closed symmetric ring structures, MAC is an asymmetric pore. This discovery has led them to believe that the shape of the pore perturbs the biophysical properties of the membrane. The team hypothesised that a subsection of the pore partially inserts into the host membrane initially to prime it for the insertion of the bulk of the complex.
Understanding the role of MAC could also potentially play a part in the fight against cancer. The functional knowledge gained from knowing the MAC's structure could ultimately be used to enhance certain existing therapies. In order to realise this aim, the team intend to continue studying the structure of MAC in even greater detail.
Related publication:
1. Serna M et al. Structural basis of complement membrane attack complex formation. Nat Commun. (2016)
doi:10.1038/ncomms10587.
XFEL Hub at Diamond
In preparation for the European XFEL (X-ray Free Electron Laser) which is due to come online in Hamburg in 2017, a centre for expertise in every aspect of XFEL experiments has been established at Diamond. Funded by the Wellcome Trust and the Biotechnology and Biological Research council (BBSRC), the UK-XFEL Hub provides support in technical development, including sample preparation, delivery systems and data analysis.
This project places the UK as the major partner of the first user consortium led project at the European XFEL which will build a dedicated instrument for serial femtosecond crystallography (SFX). The SFX Consortium is working in partnership with the European XFEL to deliver the instrument for Single Particles Clusters and Biomolecules and Serial Femtosecond Crystallography (SPB/SFX) for structure determination of single particles. The beamline will exploit two interaction regions with independent end stations; all within one, interlocked hutch. The upstream region will be commissioned first and encompasses the SPB sample environment to enable single particle imaging and serial femtosecond crystallography. The downstream interaction region (the SFX end station) will be commissioned later and will use refocused X-ray photons to support SFX methods.
Dr Allen M. Orville was hired as group leader of the XFEL Hub at Diamond in October 2015. Recruitment is underway for a beamline scientist and a postdoctoral research associate to join the team. The UK-XFEL Hub, SFX Consortium, and the SPB/SFX team will deploy several sample delivery methods first, including liquid jets, high viscosity extruders, and fixed targets. The groups will work together to support the user community in preparing for these methods through training and outreach in advance of first beam. The UK-XFEL Hub and the SFX consortium are also engaged in research and development of several novel sample delivery methods that will enable more efficient and entirely new types of experiments at XFEL and synchrotron sources.
XFEL beamtime is granted through a Peer Review proposal process ensuring that the best science is selected. The Hub at Diamond will work to beneficially impact beamtime proposals from the community, and help with experiments conducted at the LCLS (USA), SACLA (Japan), and the European XFEL, as well as other facilities as they come online.
The UK-XFEL Hub anticipates participating in SPB/SFX commissioning activities planned throughout the remainder of 2016. In October 2016 there will be a call for European XFEL “Early Science” proposals for beamtime at the SPB end station. The experiments will take place in early 2017 and provide critical opportunities for serial femtosecond crystallography. Full user operations for the two SPB/SFX end stations are anticipated to commence in 2018.


Figure 5: Dr Allen M. Orville, group leader of the XFEL Hub at Diamond.
The Hub actively supports the UK community in making full use of the transformational potential of XFELs. There will be an opportunity to highlight XFEL science and facilitate training through a workshop focused on new frontiers and emerging techniques at Diamond’s annual Synchrotron Radiation Users’ Meeting (SRUM) in September 2016. It will incorporate XFEL and cryo- EM talks in both the physical and life sciences and include a satellite workshop on day two for life science applications at the European XFEL which could potentially be put forward as part of the early science call for the SPB end station.
A working group for XFEL science is in the process of being formed and will meet regularly to help foster communication and collaboration between interested parties. As the XFEL Hub matures, the formation of a new XFEL BAG will be considered to help coordinate access between Diamond and the European XFEL, as well as potentially with the CLF and eBIC.The XFEL Hub at Diamond will help facilitate:
- serial structural biology and time-resolved experiments via sample preparation, delivery, data collection, and processing;
- the transfer of XFEL methods to synchrotron and cryo-EM sources;
- access to, and data collection from, complementary facilities such as the Diamond synchrotron, the Central Laser Facility (CLF), and the new electron Bio-Imaging Centre (eBIC).


 Diamond Annual Review 2016
Diamond Annual Review 2016

